Content from Before we Start
Last updated on 2026-02-03 | Edit this page
Estimated time: 40 minutes
Overview
Questions
- What is R and why learn it?
- How to find your way around RStudio?
- How to interact with R?
- How to install packages?
Objectives
- Navigate the RStudio interface.
- Create an R project for project analysis.
- Install additional packages using the packages tab.
- Install additional packages using R code.
This episode is adapted from Before We Start from the R for Social Scientists Carpentry lesson, licensed under a Creative Commons Attribution 4.0 License (CC BY 4.0).
What is R? What is RStudio?
R is more of a programming language than just a
statistics program. It was started by Robert Gentleman and Ross Ihaka
from the University of Auckland in 1995. They described it as “a language
for data analysis and graphics.” You can use R to create, import, and
scrape data from the web; clean and reshape it; visualize it; run
statistical analysis and modeling operations on it; text and data mine
it; and much more. The term “R” is used to refer to both
the programming language and the software that interprets the scripts
written using it.
RStudio: a user interface for R
RStudio is a user interface for working with R. It is called an Integrated Development Environment (IDE): a piece of software that provides tools to make programming easier. RStudio acts as a sort of wrapper around the R language. You can use R without RStudio, but it’s much more limiting. RStudio makes it easier to import datasets, create and write scripts, and makes using R much more effective. RStudio is also free and open source. To function correctly, RStudio needs R and therefore both need to be installed on your computer.
Why learn R?

R does not involve lots of pointing and clicking, and that’s a good thing.
The learning curve might be steeper than with other software, but with R, the results of your analysis do not rely on remembering a succession of pointing and clicking, but instead on a series of written commands, and that’s a good thing! So, if you want to redo your analysis because you collected more data, you don’t have to remember which button you clicked in which order to obtain your results; you just have to run your script again.
Working with scripts makes the steps you used in your analysis clear, and the code you write can be inspected by someone else who can give you feedback and spot mistakes. It forces you to have a deeper understanding of what you are doing, and facilitates your learning and comprehension of the methods you use.
R code is great for reproducibility.
Reproducibility is when someone else (including your future self) can obtain the same results from the same dataset when using the same analysis.
R integrates with other tools to generate manuscripts from your code. If you collect more data, or fix a mistake in your dataset, the figures and the statistical tests in your manuscript are updated automatically.
An increasing number of journals and funding agencies expect analyses to be reproducible, so knowing R will give you an edge with these requirements.
This focus on reproducibility not only helps ensure that your data adheres to the FAIR (Findable, Accessible, Interoperable, Reusable) principles but is also paving the way for software to become FAIR. As more researchers demand transparency and shareability, using R positions your code and analyses to be easily found, accessed, and reused by others.
R is interdisciplinary and extensible.
With 10,000+ packages that can be installed to extend its capabilities, R provides a framework that allows you to combine statistical approaches from many scientific disciplines to best suit the analytical framework you need to analyze your data. For instance, R has packages for image analysis, GIS, time series, population genetics, and a lot more.
R works on data of all shapes and sizes.
The skills you learn with R scale easily with the size of your dataset. Whether your dataset has hundreds or millions of lines, it won’t make much difference to you.
R is designed for data analysis. It comes with special data structures and data types that make handling of missing data and statistical factors convenient.
R can connect to spreadsheets, databases, and many other data formats, on your computer or on the web.
R produces high-quality graphics.
The plotting functionalities in R are endless, and allow you to adjust any aspect of your graph to convey most effectively the message from your data.
R has a large and welcoming community.
Thousands of people use R daily. Many of them are willing to help you through mailing lists and websites such as Stack Overflow, or on the RStudio community. Questions which are backed up with short, reproducible code snippets are more likely to attract knowledgeable responses.
Not only is R free, but it is also open-source and cross-platform.
R is also free and open source, distributed under the terms of the GNU General Public License.. This means it is free to download and use the software for any purpose, modify it, and share it. Anyone can inspect the source code to see how R works. Because of this transparency, there is less chance for mistakes, and if you (or someone else) find some, you can report and fix bugs. As a result, R users have created thousands of packages and software to enhance user experience and functionality.
Because R is open source and is supported by a large community of developers and users, there is a very large selection of third-party add-on packages which are freely available to extend R’s native capabilities.
Discussion: R for librarianship
For at least the last decade, librarians have been grappling with the ways that the “data deluge” affects our work on multiple levels–collection development, analyzing usage of the library website/space/collections, reference services, information literacy instruction, research support, accessing bibliographic metadata from third parties, and more.
Discuss some examples on how R or RStudio is useful for librarians.
- Clean messy data from the ILS & vendors
- Clean ISBNs, ISSNs, other identifiers
- Detect data errors & anomalies
- Normalize names (e.g. databases, ebooks, serials)
- Create custom subsets
- Merge and analyze data, e.g.
- Holdings and usage data from the same vendor
- Print book & ebook holdings
- COUNTER statistics
- Institutional data
- Recode variables
- Manipulate dates and times
- Create visualizations
- Provide data reference services
- Access data via APIs, including Crossref, Unpaywall, ORCID, and Sherpa-ROMeO
- Write documents to communicate findings
About RStudio
Let’s start by learning about RStudio, which is an Integrated Development Environment (IDE) for working with R.
The RStudio IDE open-source product is free under the Affero General Public License (AGPL) v3. The RStudio IDE is also available with a commercial license and priority email support from RStudio, Inc.
We will use the RStudio IDE to write code, navigate the files on our computer, inspect the variables we create, and visualize the plots we generate. RStudio can also be used for other things (e.g., version control, developing packages, writing Shiny apps) that we will not cover during the workshop.
One of the advantages of using RStudio is that all the information you need to write code is available in a single window. Additionally, RStudio provides many shortcuts, autocompletion, and highlighting for the major file types you use while developing in R. RStudio makes typing easier and less error-prone.
The RStudio Interface
Let’s take a quick tour of RStudio.
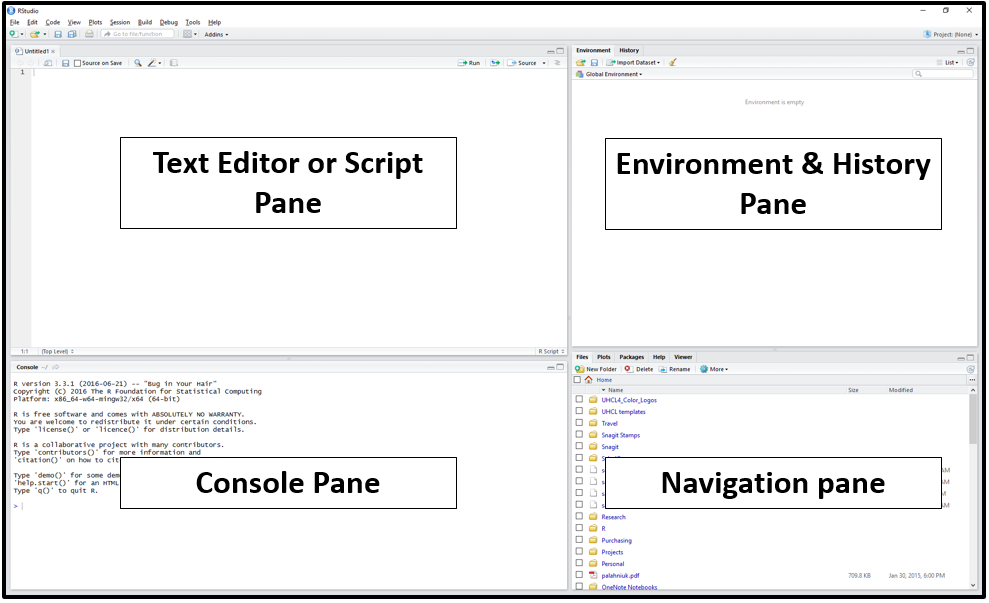
RStudio is divided into four “panes”. The placement of these panes and their content can be customized (see menu, Tools -> Global Options -> Pane Layout).
The Default Layout is:
Script Pane: This is sort of like a text editor, where you write and save a code. You can save the script as a .R file for future use and sharing, or run the code to generate an output.
Console Pane If you were just using the basic R interface, without RStudio, this is all you would see. The Console is where your scripts are executed. To execute a code, you can either run it from the Script Pane or type your code directly into the Console (you will see a blinking cursor
>prompting you to enter some code.Environment/History Pane: This will display the objects that you’ve read into what is called the “global environment.” When you read a file into R, or manually create an R object, it enters into the computer’s working memory. The History tab displays all commands that have been executed in the console.
-
Files, Plots, Packages, and Help pane This pane has multiple functions:
- Files: Navigate to files saved on your computer and in your working directory
- Plots: View plots (e.g. charts and graphs) you have created
- Packages: view add-on packages you have installed, or install new packages
- Help: Read help pages for R functions
- Viewer: View local web content
Tip: Saving your R code
To save your R codes and reuse them in the future, write and run your code from the Script Pane and save them as a .R file. This ensures your code is reproducible and that there is a complete record of what we did, and anyone (including our future selves) can easily replicate the results on their computer.
Alternatively, you can run temporary ‘test’ codes in the Console Pane – these will not be saved but are available in the History Pane.
Use the shortcuts Ctrl + 1 and Ctrl + 2 to jump between the Script and Console Panes.
Create a new project
RStudio project
An RStudio project (.Rproj) allows you to save all of the data, analyses, packages, etc. related to a specific analysis project into a single working directory.
All of the scripts within this folder can then use relative paths to files. Relative paths indicate where inside the project a file is located (as opposed to absolute paths, which point to where a file is on a specific computer).
Working this way makes it a lot easier to move your project around on your computer and share it with others without having to directly modify file paths in the individual scripts.
To create a new project, follow the instructions below.
Under the File menu, click on New project.
Select New directory, then
New project.Enter your new project name e.g. my_project. This will be your working directory for the rest of the day.
Finally, click on Create project. Create a new file where we will type our scripts. Go to
File > New File > Rscript. Click the save icon on your toolbar and save your script as “script.R”.
Interacting with R
The basis of programming is that we write down instructions for the computer to follow, and then we tell the computer to follow those instructions. We write, or code, instructions in R because it is a common language that both the computer and we can understand.
There are two main ways of interacting with R: by using the console or by using script files (plain text files that contain your code). The console pane (in RStudio, the bottom left panel) is the place where commands written in the R language can be typed and executed immediately by the computer. It is also where the results will be shown for commands that have been executed. You can type commands directly into the console and press Enter to execute those commands, but they will be forgotten when you close the session.
The prompt
The prompt (>) is the blinking
cursor in the console pane that prompts you to take action, in the
lower-left corner of R Studio. If R is ready to accept a
command, the R console shows a >
prompt. If R receives a command (by typing, copy-pasting, or sent from
the script editor using Ctrl + Enter), R will try
to execute it and, when ready, will show the results and come back with
a new > prompt to wait for new commands. We type
commands into the prompt, and press the Enter key to
evaluate (also called execute or run) those
commands.
You can use R like a calculator:
R
2 + 2 # Type 2 + 2 in the console to run the command
Tip: Moving your cursor
In the console, you can press the up and down keys on your keyboard to cycle through previously executed commands.
Because we want our code and workflow to be reproducible, it is better to type the commands we want in the script editor and save the script. This way, there is a complete record of what we did, and anyone (including our future selves!) can easily replicate the results on their computer.
RStudio allows you to execute commands directly from the script editor by using the Ctrl + Enter shortcut (on Mac, Cmd + Return will work).
The command on the current line in the script (indicated by the cursor) or all of the commands in selected text will be sent to the console and executed when you press Ctrl + Enter. If there is information in the console you do not need anymore, you can clear it with Ctrl + L.
You can find other keyboard shortcuts in this RStudio cheatsheet about the RStudio IDE.
Tip: Incomplete commands
If R recognises that a command is incomplete, a + prompt
will appear in the console. This means that R is still waiting for you
to enter more text.
Usually, this occurs because you have not ‘closed’ a parenthesis or
quotation. When this happens, click on the console and press
Esc; this will cancel the incomplete command and return you
to the > prompt. You can then proofread the command(s)
you entered and correct the error.
Installing R packages
When you download R it already has a number of functions built in: these encompass what is called Base R. However, many R users write their own libraries of functions, package them together in R Packages, and provide them to the R community at no charge. Some examples include the dplyr and ggplot2 packages, which we will learn more in the coming chapters.
The CRAN repository
The Comprehensive R Archive Network (CRAN) is the main repository for R packages, and that organization maintains strict standards in order for a package to be listed–for example, it must include clear descriptions of the functions, and it must not track or tamper with the user’s R session.
See this page from RStudio for a good list of useful R packages. In addition to CRAN, R users can make their code and packages available from GitHub or Bioconductor (for computational biology and bioinformatics).
Installing packages using the Packages tab
Installing CRAN packages can be done from the Packages tab
in the Files, Plots, Packages, and Help pane. Click Install
and type in the name of the package you’re looking for.
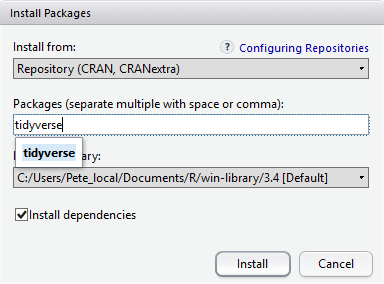
At the bottom of the Install Packages window is a check box to ‘Install’ dependencies. This is ticked by default, which is usually what you want. Packages can (and do) make use of functionality built into other packages, so for the functionality contained in the package you are installing to work properly, there may be other packages which have to be installed with them. The ‘Install dependencies’ option makes sure that this happens.
Exercise
Use the install option from the packages tab to install the ‘tidyverse’ package.
From the packages tab, click ‘Install’ from the toolbar and type ‘tidyverse’ into the textbox, then click ‘install’. The ‘tidyverse’ package is really a package of packages, including ‘ggplot2’ and ‘dplyr’, both of which require other packages to run correctly. All of these packages will be installed automatically.
Depending on what packages have previously been installed in your R environment, the install of ‘tidyverse’ could be very quick or could take several minutes. As the install proceeds, messages relating to its progress will be written to the console. You will be able to see all of the packages which are actually being installed.
Installing packages using R code
If you were watching the console window when you started the install of ‘tidyverse’, you may have noticed that the line
R
install.packages("tidyverse")
was written to the console before the start of the installation messages.
You could also have installed the
tidyverse packages by running this command
directly in the R console. Run help(install.packages) to
learn more about how to do it this way.
Because the install process accesses the CRAN repository, you will need an Internet connection to install packages.
It is also possible to install packages from other repositories, as well as Github or the local file system, but we won’t be looking at these options in this lesson.
R Resources
Learning R
swirlis a package you can install in R to learn about R and data science interactively. Just typeinstall.packages("swirl")into your R console, load the package by typinglibrary("swirl"), and then typeswirl(). Read more at swirl.Try R is a browser-based interactive tutorial developed by Code School.
Anthony Damico’s twotorials are a series of 2 minute videos demonstrating several basic tasks in R.
Cookbook for R by Winston Change provides solutions to common tasks and problems in analyzing data.
If you’re up for a challenge, try the free R Programming MOOC in Coursera by Roger Peng.
Books:
- R For Data Science by Garrett Grolemund & Hadley Wickham [free]
- An Introduction to Data Cleaning with R by Edwin de Jonge & Mark van der Loo [free]
- YaRrr! The Pirate’s Guide to R by Nathaniel D. Phillips [free]
- Springer’s Use R! series [not free] is mostly specialized, but it has some excellent introductions including Alain F. Zuur et al.’s A Beginner’s Guide to R and Phil Spector’s Data Manipulation in R.
- Other Carpentries R lesson:
- SWC – Programming with R
- SWC – R for Reproducible Scientific Analysis
- LC - Introduction to R and litsearchr (pre-alpha)
- Data Analysis and Visualization in R for Ecologists
- Introduction to R and RStudio for Genomics
Datasets
If you need some data to play with, type data() in the
console for a list of data sets. To load a dataset, type it like this:
data(mtcars). Type help(mtcars) to learn more
about it. You can then perform operations, e.g.
R
head(mtcars)
nrow(mtcars)
mean(mtcars$mpg)
sixCylinder <- mtcars[mtcars$cyl == 6, ]
See also rdatamining.com’s list of free datasets.
Cheat Sheets
List of R Cheat Sheets:
- Base R Cheat Sheet by Mhairi McNeill
- Data Transformation with dplyr Cheat Sheet by RStudio
- Data Wrangling with dplyr and tidyr Cheat Sheet by RStudio
- Complete list of RStudio cheatsheets
You can find more cheat sheets in RStudio by going to the
Help panel then clicking on the
Style guides
Use these resources to write cleaner code, according to established style conventions
- Hadley Wickham’s Style Guide
- Google’s R Style Guide
- Tip: highlight code in your script pane and press Ctrl/Cmd + I on your keyboard to automatically fix the indents
Credit
Parts of this episode have been inspired by the following:
- “Before We Start” R for Social Scientists Carpentry Lesson. CC BY 4.0.
- Roger Peng’s Computing for Data Analysis videos
- Lisa Federer’s Introduction to R for Non-Programmers
- Brad Boehmke’s Intro to R Bootcamp
- Navigate round RStudio and create an
Rprojfile. - Use RStudio to write and run R programs.
- Install packages using the Packages tab or the
install.packages()command.
Content from Introduction to R
Last updated on 2026-02-19 | Edit this page
Estimated time: 80 minutes
Overview
Questions
- What is an object?
- What is a function and how can we pass arguments to functions?
- How can values be initially assigned to variables of different data types?
- What are the different data types in R?
- How can a vector be created What are the available data types?
- How can subsets be extracted from vectors?
- How does R treat missing values?
- How can we deal with missing values in R?
Objectives
- Assign values to objects in R.
- Learn how to name objects.
- Use comments to inform script.
- Solve simple arithmetic operations in R.
- Call functions and use arguments to change their default options.
- Where to get help with R functions and packages.
- Recognise the different data types in R.
- Create, inspect and manipulate the contents of a vector.
- Subset and extract values from vectors.
- Analyze vectors with missing data.
Creating objects in R
Objects vs. variables
What are known as objects in R are known as
variables in many other programming languages. Depending on
the context, object and variable can have
drastically different meanings. However, in this lesson, the two words
are used synonymously. For more information see: https://cran.r-project.org/doc/manuals/r-release/R-lang.html#Objects
We will start by typing a simple mathematical operation in the console:
R
3 + 5
OUTPUT
[1] 8R
7 * 2 # multiply 7 by 2
OUTPUT
[1] 14R
sqrt(36) # take the square root of 36
OUTPUT
[1] 6However, to do useful and interesting things, we need to assign values to objects.
To create an object, we need to provide: - a name (e.g. ‘first_number’) - a value (e.g. ‘1’) - the assignment operator (‘<-’)
R
time_minutes <- 5 # assign the number 5 to the object time_minutes
The assignment operator
<-
<- is the assignment operator. It assigns values on
the right to objects on the left. Here we are creating a symbol called
time_minutes and assigning it the numeric value 5. Some R
users would say “time_minutes gets 5.”
time_minutes is now a numeric vector with one
element.
The ‘=’ works too, but is most commonly used in passing arguments to functions (more on functions later). There are shortcuts for the R assignment operator: - Windows shortcut: Alt+- - Mac shortcut: Option+-
When you assign something to a symbol, nothing happens in the
console, but in the Environment pane in the upper right, you will notice
a new object, time_minutes.
Tips on naming objects
Here are some tips for naming objects in R:
-
Do not use names of functions that already exist in
R: There are some fundamental functions in R (e.g.,
if,else,for, see the list of reserved words for a complete list. In general, even if it’s allowed, it’s best to not use other function names (e.g.,c,T,mean,data,df,weights). If in doubt, check the help to see if the name is already in use. -
R is case sensitive:
ageis different fromAgeandyis different fromY. -
No blank spaces or symbols other than underscores:
R users get around this in a couple of ways, either through
capitalization (e.g.
myData) or underscores (e.g.my_data). It’s also best to avoid dots (.) within an object name as inmy.dataset. There are many functions in R with dots in their names for historical reasons, but dots have a special meaning in R (for methods) and other programming languages. -
Do not begin with numbers or symbols:
2xis not valid, butx2is. -
Be descriptive, but make your variable names short:
It’s good practice to be descriptive with your variable names. If you’re
loading in a lot of data, choosing
myDataorxas a name may not be as helpful as, say,ebookUsage. Finally, keep your variable names short, since you will likely be typing them in frequently.
Evaluating Expressions
If you now type time_minutes into the console, and press
Enter on your keyboard, R will evaluate the expression. In this
case, R will print the elements that are assigned to
time_minutes (the number 5). We can do this easily since y
only has one element, but if you do this with a large dataset loaded
into R, it will overload your console because it will print the entire
thing. The [1] indicates that the number 5 is the first
element of this vector.
When assigning a value to an object, R does not print anything to the console. You can force R to print the value by using parentheses or by typing the object name:
R
time_minutes <- 5 # doesn't print anything
(time_minutes <- 5) # putting parenthesis around the call prints the value of y
OUTPUT
[1] 5R
time_minutes # so does typing the name of the object
OUTPUT
[1] 5R
print(time_minutes) # so does using the print() function.
OUTPUT
[1] 5Now that R has time_minutes in memory, we can do
arithmetic with it. For instance, we may want to convert it into seconds
(60 seconds in 1 minute):
R
60 * time_minutes
OUTPUT
[1] 300We can also change an object’s value by assigning it a new one:
R
time_minutes <- 10
60 * time_minutes
OUTPUT
[1] 600This overwrites the previous value without prompting you, so be
careful! Also, assigning a value to one object does not change the
values of other objects For example, let’s store the time in seconds in
a new object, time_seconds:
R
time_seconds <- 60 * time_minutes
Then change time_minutes to 30:
R
time_minutes <- 30
Exercise
What do you think is the current content of the object
time_seconds? 600 or 1800?
The value of time_seconds is still 600 because you have
not re-run the line time_seconds <- 60 * time_minutes
since changing the value of time_minutes.
Removing objects from the environment
To remove an object from your R environment, use the
rm() function. Remove multiple objects with
rm(list = c("add", "objects", "here)), adding the objects
in c() using quotation marks.
To remove all objects, use rm(list = ls()) or click the
broom icon in the Environment Pane, next to “Import Dataset.”
R
x <- 5
y <- 10
z <- 15
rm(x) # remove x
rm(list =c("y", "z")) # remove y and z
rm(list = ls()) # remove all objects
Tip: Use whitespace for readability
The white spaces surrounding the assignment operator
<- are unnecessary. However, including them does make
your code easier to read. There are several style guides you can follow,
and choosing one is up to you, but consistency is key!
A style guide we recommend is the Tidyverse style guide.
Exercise
Create two variables my_length and my_width
and assign them any numeric values you want. Create a third variable
my_area and give it a value based on the the multiplication
of my_length and my_width. Show that changing
the values of either my_length and my_width
does not affect the value of my_area.
R
my_length <- 2.5
my_width <- 3.2
my_area <- my_length * my_width
area
ERROR
Error:
! object 'area' not foundR
# change the values of my_length and my_width
my_length <- 7.0
my_width <- 6.5
# the value of my_area isn't changed
my_area
OUTPUT
[1] 8Comments
All programming languages allow the programmer to include comments in
their code. To do this in R we use the # character.
Anything to the right of the # sign and up to the end of
the line is treated as a comment and will not be evaluated by R. You can
start lines with comments or include them after any code on the
line.
Comments are essential to helping you remember what your code does, and explaining it to others. Commenting code, along with documenting how data is collected and explaining what each variable represents, is essential to reproducible research. See the Software Carpentry lesson on R for Reproducible Scientific Analysis.
R
time_minutes <- 5 # time in minutes
time_seconds <- 60 * time_minutes # convert to seconds
time_seconds # print time in seconds
OUTPUT
[1] 300RStudio makes it easy to comment or uncomment a paragraph: after selecting the lines you want to comment, press at the same time on your keyboard Ctrl + Shift + C. If you only want to comment out one line, you can put the cursor at any location of that line (i.e. no need to select the whole line), then press Ctrl + Shift + C.
Functions and their arguments
R is a “functional programming language,” meaning it contains a number of functions you use to do something with your data. Functions are “canned scripts” that automate more complicated sets of commands. Many functions are predefined, or can be made available by importing R packages as we saw in the “Before We Start” lesson.
Call a function on a variable by entering the function into
the console, followed by parentheses and the variables. A function
usually gets one or more inputs called arguments. For example,
if you want to take the sum of 3 and 4, you can type in
sum(3, 4). In this case, the arguments must be a number,
and the return value (the output) is the sum of those numbers. An
example of a function call is:
R
sum(3, 4)
The function is.function() will check if an argument is
a function in R. If it is a function, it will print TRUE to
the console.
Functions can be nested within each other. For example,
sqrt() takes the square root of the number provided in the
function call. Therefore you can run sum(sqrt(9), 4) to
take the sum of the square root of 9 and add it to 4.
Typing a question mark before a function will pull the help page up
in the Files, Plots, Packages, and Help pane in the lower right. Type
?sum to view the help page for the sum
function. You can also call help(sum). This will provide
the description of the function, how it is to be used, and the
arguments.
In the case of sum(), the ellipses . . .
represent an unlimited number of numeric elements.
R
is.function(sum) # check to see if sum() is a function
sum(3, 4, 5, 6, 7) # sum takes an unlimited number (. . .) of numeric elements
Arguments
Some functions take arguments which may either be specified by the user, or, if left out, take on a default value. However, if you want something specific, you can specify a value of your choice which will be used instead of the default. This is called passing an argument to the function.
For example, sum() takes the argument option
na.rm. If you check the help page for sum (call
?sum), you can see that na.rm requires a
logical (TRUE/FALSE) value specifying whether
NA values (missing data) should be removed when the
argument is evaluated.
By default, na.rm is set to FALSE, so
evaluating a sum with missing values will return NA:
R
sum(3, 4, NA) #
OUTPUT
[1] NAEven though we do not see the argument here, it is operating in the
background, as the NA value remains. 3 + 4 +
NA is NA.
But setting the argument na.rm to TRUE will
remove the NA:
R
sum(3, 4, NA, na.rm = TRUE)
OUTPUT
[1] 7It is very important to understand the different arguments that
functions take, the values that can be added to those functions, and the
default arguments. Arguments can be anything, not only TRUE
or FALSE, but also other objects. Exactly what each
argument means differs per function, and must be looked up in the
documentation.
It’s good practice to put the non-optional arguments first in your function call, and to specify the names of all optional arguments. If you don’t, someone reading your code might have to look up the definition of a function with unfamiliar arguments to understand what you’re doing.
Getting help
You can check the current version of R by running:
R
sessionInfo()
OUTPUT
R version 4.5.2 (2025-10-31)
Platform: x86_64-pc-linux-gnu
Running under: Ubuntu 24.04.3 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C
[3] LC_TIME=en_US.UTF-8 LC_COLLATE=en_US.UTF-8
[5] LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C
[9] LC_ADDRESS=C LC_TELEPHONE=C
[11] LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
time zone: Etc/UTC
tzcode source: system (glibc)
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] compiler_4.5.2 cli_3.6.5 tools_4.5.2 pillar_1.11.1
[5] otel_0.2.0 glue_1.8.0 yaml_2.3.12 vctrs_0.7.0
[9] knitr_1.51 xfun_0.56 lifecycle_1.0.5 rlang_1.1.7
[13] renv_1.1.6 evaluate_1.0.5 R version 3.2.3 (2015-12-10)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 14.04.3 LTS
locale:
[1] LC_CTYPE=en_US.UTF-8 LC_NUMERIC=C LC_TIME=en_US.UTF-8
[4] LC_COLLATE=en_US.UTF-8 LC_MONETARY=en_US.UTF-8 LC_MESSAGES=en_US.UTF-8
[7] LC_PAPER=en_US.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=en_US.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
loaded via a namespace (and not attached):
[1] tools_3.2.3 packrat_0.4.9-1Using the ? for Help files
R provide help files for functions. The general syntax to search for help on any function is the ? or help() command:
R
?function_name
OUTPUT
No documentation for 'function_name' in specified packages and libraries:
you could try '??function_name'R
help(function_name)
OUTPUT
No documentation for 'function_name' in specified packages and libraries:
you could try '??function_name'For example, take a look at the ?summary() function.
R
?summary()
This will load up a Help page on the bottom right panel in RStudio.
Each help page is broken down into sections:
- Description: An extended description of what the function does.
- Usage: The arguments of the function and their default values (which can be changed).
- Arguments: An explanation of the data each argument is expecting.
- Details: Any important details to be aware of.
- Value: The data the function returns.
- See Also: Any related functions you might find useful.
- Examples: Some examples for how to use the function.
You can also call out the Help file for
special operators.
R
?"<-"
?`<-`
Tip: Use TAB to see a list of arguments
After typing out a function, you can call out a list of available arguments for that particular function by pressing the TAB key within the function brackets.
Tip: Package vignettes
Many packages come with “vignettes”–these are tutorials and extended documentation. Without any arguments, vignette() will list all vignettes for all installed packages; vignette(package=“package-name”) will list all available vignettes for package-name, and vignette(“vignette-name”) will open the specified vignette.
Using the ?? for a fuzzy search
If you’re not sure what package a function is in or how it’s specifically spelled, you can do a fuzzy search.
R
??function_name
OUTPUT
No vignettes or demos or help files found with alias or concept or
title matching 'function_name' using regular expression matching.A fuzzy search is when you search for an approximate string match. For example, you may want to look for a function that performs the chi-squared test:
R
??chisquare
Discussion
Type in ?round at the console and then look at the
output in the Help pane. What other functions exist that are similar to
round? How do you use the digits parameter in
the round function?
Other ways to get help
As mentioned above, you can use a question mark ? to
know more about a function (for example, typing ?round).
However, there are several other ways that people often get help when
they are stuck with their R code.
- Search the internet: paste the last line of your error message or
“R” and a short description of what you want to do into your favourite
search engine and you will usually find several examples where other
people have encountered the same problem and came looking for help.
- Stack Overflow can be particularly helpful for this: answers to
questions are presented as a ranked thread ordered according to how
useful other users found them to be. You can search using the
[r]tag. - Take care: copying and pasting code written by somebody else is risky unless you understand exactly what it is doing!
- Stack Overflow can be particularly helpful for this: answers to
questions are presented as a ranked thread ordered according to how
useful other users found them to be. You can search using the
- Ask somebody “in the real world”. If you have a colleague or friend with more expertise in R than you have, show them the problem you are having and ask them for help.
- Sometimes, the act of articulating your question can help you to identify what is going wrong. This is known as “rubber duck debugging” among programmers.
Generative AI
The section on generative AI is intended to be concise but Instructors may choose to devote more time to the topic in a workshop. Depending on your own level of experience and comfort with talking about and using these tools, you could choose to do any of the following:
- Explain how large language models work and are trained, and/or the difference between generative AI, other forms of AI that currently exist, and the limits of what LLMs can do (e.g., they can’t “reason”).
- Demonstrate how you recommend that learners use generative AI.
- Discuss the ethical concerns listed below, as well as others that you are aware of, to help learners make an informed choice about whether or not to use generative AI tools.
This is a fast-moving technology. If you are preparing to teach this section and you feel it has become outdated, please open an issue on the lesson repository to let the Maintainers know and/or a pull request to suggest updates and improvements.
It is increasingly common for people to use generative AI chatbots such as ChatGPT to get help while coding. You will probably receive some useful guidance by presenting your error message to the chatbot and asking it what went wrong.
However, the way this help is provided by the chatbot is different. Answers on Stack Overflow have (probably) been given by a human as a direct response to the question asked. But generative AI chatbots, which are based on advanced statistical models called Large Language Models (or LLMs), respond by generating the most likely sequence of text that would follow the prompt they are given.
While responses from generative AI tools can often be helpful, they are not always reliable. These tools sometimes generate plausible but incorrect or misleading information, so (just as with an answer found on the internet) it is essential to verify their accuracy. You need the knowledge and skills to be able to understand these responses, to judge whether or not they are accurate, and to fix any errors in the code it offers you.
In addition to asking for help, programmers can use generative AI tools to generate code from scratch; extend, improve and reorganize existing code; translate code between programming languages; figure out what terms to use in a search of the internet; and more. However, there are drawbacks that you should be aware of.
The models used by these tools have been “trained” on very large volumes of data, much of it taken from the internet, and the responses they produce reflect that training data, and may recapitulate its inaccuracies or biases. The environmental costs (energy and water use) of LLMs are a lot higher than other technologies, both during development (known as training) and when an individual user uses one (also called inference). For more information see the AI Environmental Impact Primer developed by researchers at HuggingFace, an AI hosting platform. Concerns also exist about the way the data for this training was obtained, with questions raised about whether the people developing the LLMs had permission to use it. Other ethical concerns have also been raised, such as reports that workers were exploited during the training process.
We recommend that you avoid getting help from generative AI during the workshop for several reasons:
- For most problems you will encounter at this stage, help and answers can be found among the first results returned by searching the internet.
- The foundational knowledge and skills you will learn in this lesson by writing and fixing your own programs are essential to be able to evaluate the correctness and safety of any code you receive from online help or a generative AI chatbot. If you choose to use these tools in the future, the expertise you gain from learning and practicing these fundamentals on your own will help you use them more effectively.
- As you start out with programming, the mistakes you make will be the kinds that have also been made – and overcome! – by everybody else who learned to program before you. Since these mistakes and the questions you are likely to have at this stage are common, they are also better represented than other, more specialized problems and tasks in the data that was used to train generative AI tools. This means that a generative AI chatbot is more likely to produce accurate responses to questions that novices ask, which could give you a false impression of how reliable they will be when you are ready to do things that are more advanced.
Vectors and data types
A vector is the most common and basic data type in R, and is pretty
much the workhorse of R. A vector is a sequence of elements of the same
type. Vectors can only contain “homogenous” data–in other
words, all data must be of the same type. The type of a vector
determines what kind of analysis you can do on it. For example, you can
perform mathematical operations on numeric objects, but not
on character objects.
We can assign a series of values to a vector using the
c() function. c() stands for combine. If you
read the help files for c() by calling
help(c), you can see that it takes an unlimited
. . . number of arguments.
For example we can create a vector of checkouts for a collection of
books and assign it to a new object checkouts:
R
checkouts <- c(25, 15, 18)
checkouts
OUTPUT
[1] 25 15 18A vector can also contain characters. For example, we can have a
vector of the book titles (title) and authors
(author):
R
title <- c("Macbeth","Dracula","1984")
Tip: Using quotes
The quotes around “Macbeth”, etc. are essential here. Without the
quotes R will assume there are objects called Macbeth and
Dracula in the environment. As these objects don’t yet
exist in R’s memory, there will be an error message.
Inspecting a vector
There are many functions that allow you to inspect the content of a vector.
For instance, length() tells you how many elements are
in a particular vector:
R
length(checkouts) # print the number of values in the checkouts vector
OUTPUT
[1] 3An important feature of a vector, is that all of the elements are the
same type of data. The function class() indicates the class
(the type of element) of an object:
R
class(checkouts)
OUTPUT
[1] "numeric"R
class(title)
OUTPUT
[1] "character"Type ?str into the console to read the description of
the str function. You can call str() on an R
object to compactly display information about it, including the data
type, the number of elements, and a printout of the first few
elements.
R
str(checkouts)
OUTPUT
num [1:3] 25 15 18R
str(title)
OUTPUT
chr [1:3] "Macbeth" "Dracula" "1984"You can use the c() function to add other elements to
your vector:
R
author <- "Stoker"
author <- c(author, "Orwell") # add to the end of the vector
author <- c("Shakespeare", author)
author
OUTPUT
[1] "Shakespeare" "Stoker" "Orwell" Reminder
To know what a function does, type ?function_name into
the console. For example, you can type ?str to read the
description for the str() function.
In the first line, we create a character vector author
with a single value "Stoker". In the second line, we add
the value "Orwell" to it, and save the result back into
author. Then we add the value "Shakespeare" to
the beginning, again saving the result back into
author.
We can do this over and over again to grow a vector, or assemble a dataset. As we program, this may be useful to add results that we are collecting or calculating.
Vectors are one of the many data structures that R
uses. Other important ones are lists (list), matrices
(matrix), data frames (data.frame), factors
(factor) and arrays (array).
Atomic vectors
An atomic vector is the simplest R data
type and is a linear vector of a single type. Above, we saw 2
of the 6 main atomic vector types that R uses:
"character" and "numeric" (or
"double"). These are the basic building blocks that all R
objects are built from. The other 4 atomic vector types
are:
-
"logical"forTRUEandFALSE(the boolean data type) -
"integer"for integer numbers (e.g.,2L, theLindicates to R that it’s an integer) -
"complex"to represent complex numbers with real and imaginary parts (e.g.,1 + 4i) and that’s all we’re going to say about them -
"raw"for bitstreams that we won’t discuss further
You can check the type of your vector using the typeof()
function and inputting your vector as the argument.
Exercise
We’ve seen that atomic vectors can be of type character, numeric (or double), integer, and logical. But what happens if we try to mix these types in a single vector?
R implicitly converts them to all be the same type.
Exercise (continued)
What will happen in each of these examples? (hint: use
typeof() to check the data type of your objects):
R
num_char <- c(1, 2, 3, "a")
num_logical <- c(1, 2, 3, TRUE)
char_logical <- c("a", "b", "c", TRUE)
tricky <- c(1, 2, 3, "4")
Why do you think it happens?
Vectors can be of only one data type. R tries to convert (coerce) the content of this vector to find a “common denominator” that doesn’t lose any information.
Exercise (continued)
How many values in combined_logical are
"TRUE" (as a character) in the following example:
R
num_logical <- c(1, 2, 3, TRUE)
char_logical <- c("a", "b", "c", TRUE)
combined_logical <- c(num_logical, char_logical)
Only one. There is no memory of past data types, and the coercion
happens the first time the vector is evaluated. Therefore, the
TRUE in num_logical gets converted into a
1 before it gets converted into "1" in
combined_logical.
Coercion
You’ve probably noticed that objects of different types get converted into a single, shared type within a vector. In R, we call converting objects from one class into another class coercion. These conversions happen according to a hierarchy, whereby some types get preferentially coerced into other types. This hierarchy is: logical < integer < numeric < complex < character < list.
You can also coerce a vector to be a specific data type with
as.character(), as.logical(),
as.numeric(), etc. For example, to coerce a number to a
character:
R
x <- as.character(200)
We can test this in a few ways: if we print x to the
console, we see quotation marks around it, letting us know it is a
character:
R
x
OUTPUT
[1] "200"We can also call class()
R
class(x)
OUTPUT
[1] "character"And if we try to add a number to x, we will get an error
message non-numeric argument to binary operator--in other
words, x is non-numeric and cannot be added to a
number.
R
x + 5
Subsetting vectors
If we want to subset (or extract) one or several values from a
vector, we must provide one or several indices in square brackets. For
this example, we will use the state data, which is built
into R and includes data related to the 50 states of the U.S.A. Type
?state to see the included datasets.
state.name is a built in vector in R of all U.S.
states:
R
state.name
OUTPUT
[1] "Alabama" "Alaska" "Arizona" "Arkansas"
[5] "California" "Colorado" "Connecticut" "Delaware"
[9] "Florida" "Georgia" "Hawaii" "Idaho"
[13] "Illinois" "Indiana" "Iowa" "Kansas"
[17] "Kentucky" "Louisiana" "Maine" "Maryland"
[21] "Massachusetts" "Michigan" "Minnesota" "Mississippi"
[25] "Missouri" "Montana" "Nebraska" "Nevada"
[29] "New Hampshire" "New Jersey" "New Mexico" "New York"
[33] "North Carolina" "North Dakota" "Ohio" "Oklahoma"
[37] "Oregon" "Pennsylvania" "Rhode Island" "South Carolina"
[41] "South Dakota" "Tennessee" "Texas" "Utah"
[45] "Vermont" "Virginia" "Washington" "West Virginia"
[49] "Wisconsin" "Wyoming" R
state.name[1]
OUTPUT
[1] "Alabama"You can use the : colon to create a vector of
consecutive numbers.
R
state.name[1:5]
OUTPUT
[1] "Alabama" "Alaska" "Arizona" "Arkansas" "California"If the numbers are not consecutive, you must use the c()
function:
R
state.name[c(1, 10, 20)]
OUTPUT
[1] "Alabama" "Georgia" "Maryland"We can also repeat the indices to create an object with more elements than the original one:
R
state.name[c(1, 2, 3, 2, 1, 3)]
OUTPUT
[1] "Alabama" "Alaska" "Arizona" "Alaska" "Alabama" "Arizona"R indices start at 1. Programming languages like Fortran, MATLAB, Julia, and R start counting at 1, because that’s what human beings typically do. Languages in the C family (including C++, Java, Perl, and Python) count from 0 because that’s simpler for computers to do.
Conditional subsetting
Another common way of subsetting is by using a logical vector.
TRUE will select the element with the same index, while
FALSE will not:
R
five_states <- state.name[1:5]
five_states[c(TRUE, FALSE, TRUE, FALSE, TRUE)]
OUTPUT
[1] "Alabama" "Arizona" "California"Typically, these logical vectors are not typed by hand, but are the
output of other functions or logical tests. state.area is a
vector of state areas in square miles. We can use the <
operator to return a logical vector with TRUE for the indices that meet
the condition:
R
state.area < 10000
OUTPUT
[1] FALSE FALSE FALSE FALSE FALSE FALSE TRUE TRUE FALSE FALSE TRUE FALSE
[13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE
[25] FALSE FALSE FALSE FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
[37] FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE
[49] FALSE FALSER
state.area[state.area < 10000]
OUTPUT
[1] 5009 2057 6450 8257 9304 7836 1214 9609The first expression gives us a logical vector of length 50, where
TRUE represents those states with areas less than 10,000
square miles. The second expression subsets state.name to
include only those names where the value is TRUE.
You can also specify character values. state.region
gives the region that each state belongs to:
R
state.region == "Northeast"
OUTPUT
[1] FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE
[13] FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE TRUE FALSE FALSE FALSE
[25] FALSE FALSE FALSE FALSE TRUE TRUE FALSE TRUE FALSE FALSE FALSE FALSE
[37] FALSE TRUE TRUE FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE
[49] FALSE FALSER
state.name[state.region == "Northeast"]
OUTPUT
[1] "Connecticut" "Maine" "Massachusetts" "New Hampshire"
[5] "New Jersey" "New York" "Pennsylvania" "Rhode Island"
[9] "Vermont" Again, a TRUE/FALSE index of all 50 states where the
region is the Northeast, followed by a subset of state.name
to return only those TRUE values.
Sometimes you need to do multiple logical tests (think Boolean
logic). You can combine multiple tests using | (at least
one of the conditions is true, OR) or & (both
conditions are true, AND). Use help(Logic) to read the help
file.
R
state.name[state.area < 10000 | state.region == "Northeast"]
OUTPUT
[1] "Connecticut" "Delaware" "Hawaii" "Maine"
[5] "Massachusetts" "New Hampshire" "New Jersey" "New York"
[9] "Pennsylvania" "Rhode Island" "Vermont" R
state.name[state.area < 10000 & state.region == "Northeast"]
OUTPUT
[1] "Connecticut" "Massachusetts" "New Hampshire" "New Jersey"
[5] "Rhode Island" "Vermont" The first result includes both states with fewer than 10,000 sq. mi. and all states in the Northeast. New York, Pennsylvania, Delaware and Maine have areas with greater than 10,000 square miles, but are in the Northeastern U.S. Hawaii is not in the Northeast, but it has fewer than 10,000 square miles. The second result includes only states that are in the Northeast and have fewer than 10,000 sq. mi.
R contains a number of operators you can use to compare values. Use
help(Comparison) to read the R help file. Note that
two equal signs (==) are used for
evaluating equality (because one equals sign (=) is used
for assigning variables).
A common task is to search for certain strings in a vector. One could
use the “or” operator | to test for equality to multiple
values, but this can quickly become tedious. The function
%in% allows you to test if any of the elements of a search
vector are found:
R
west_coast <- c("California", "Oregon", "Washington")
state.name[state.name == "California" | state.name == "Oregon" | state.name == "Washington"]
OUTPUT
[1] "California" "Oregon" "Washington"R
state.name %in% west_coast
OUTPUT
[1] FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[25] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
[37] TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE
[49] FALSE FALSER
state.name[state.name %in% west_coast]
OUTPUT
[1] "California" "Oregon" "Washington"Missing data
As R was designed to analyze datasets, it includes the concept of
missing data (which is uncommon in other programming languages). Missing
data are represented in vectors as NA. R functions have
special actions when they encounter NA.
When doing operations on numbers, most functions will return
NA if the data you are working with include missing values.
This feature makes it harder to overlook the cases where you are dealing
with missing data. As we saw above, you can add the argument
na.rm=TRUE to calculate the result while ignoring the
missing values.
R
rooms <- c(2, 1, 1, NA, 4)
mean(rooms)
OUTPUT
[1] NAR
max(rooms)
OUTPUT
[1] NAR
mean(rooms, na.rm = TRUE)
OUTPUT
[1] 2R
max(rooms, na.rm = TRUE)
OUTPUT
[1] 4If your data include missing values, you may want to become familiar
with the functions is.na(), na.omit(), and
complete.cases(). See below for examples.
R
## Use any() to check if any values are missing
any(is.na(rooms))
OUTPUT
[1] TRUER
## Use table() to tell you how many are missing vs. not missing
table(is.na(rooms))
OUTPUT
FALSE TRUE
4 1 R
## Identify those elements that are not missing values.
complete.cases(rooms)
OUTPUT
[1] TRUE TRUE TRUE FALSE TRUER
## Identify those elements that are missing values.
is.na(rooms)
OUTPUT
[1] FALSE FALSE FALSE TRUE FALSER
## Extract those elements that are not missing values.
rooms[complete.cases(rooms)]
OUTPUT
[1] 2 1 1 4You can also use !is.na(rooms), which is exactly the
same as complete.cases(rooms). The exclamation mark
indicates logical negation.
R
!c(TRUE, FALSE)
OUTPUT
[1] FALSE TRUEHow you deal with missing data in your analysis is a decision you will have to make–do you remove it entirely? Do you replace it with zeros? That will depend on your own methodological questions.
Exercise
- Using this vector of rooms, create a new vector with the NAs removed.
R
rooms <- c(1, 2, 1, 1, NA, 3, 1, 3, 2, 1, 1, 8, 3, 1, NA, 1)
Use the function
median()to calculate the median of theroomsvector.Use R to figure out how many households in the room variable have more than 2 rooms.
R
rooms <- c(1, 2, 1, 1, NA, 3, 1, 3, 2, 1, 1, 8, 3, 1, NA, 1)
rooms_no_na <- rooms[!is.na(rooms)]
# or
rooms_no_na <- na.omit(rooms)
# 2.
median(rooms, na.rm = TRUE)
OUTPUT
[1] 1R
# 3.
rooms_above_2 <- rooms_no_na[rooms_no_na > 2]
length(rooms_above_2)
OUTPUT
[1] 4Now that we have learned how to write scripts, and the basics of R’s data structures, we are ready to start working with the library catalog dataset and learn about data frames.
- Assign values to object using the assignment operator <-. Remove
existing objects using the
rm()function. - Add comments in R scripts using the
#operator. - Define and use R functions and arguments.
- Getting help with the
?,??andhelp()functions. - Define the following terms as they relate to R: object, vector, assign, call, function.
- Create or add new objects to a vector using the c() function. Subset
vectors using
[]. - Deal with missing data in vectors using the is.na(), na.omit(), and complete.cases() functions.
Content from Starting with Data
Last updated on 2026-02-03 | Edit this page
Estimated time: 80 minutes
Overview
Questions
- What is a working directory?
- How can I create new sub-directories in R?
- How can I read a complete csv file into R?
- How can I get basic summary information about my dataset?
- How can I extract certain rows and columns from my data frame?
- How can I deal with missing values in R?
Objectives
- Set up the working directory and sub-directories.
- Load external data from a .csv file into a data frame.
- Describe what a data frame is.
- Summarize the contents of a data frame.
- Indexing and subsetting data frames.
- Explore missing values in data frames.
- Use logical operators in data frames.
Create a new R project
Let’s create a new project library_carpentry.Rproj in
RStudio to play with our dataset.
Reminder
In the previous episode Before We Start,
we briefly walk through how to create an R project to keep all your
scripts, data and analysis. If you need a refreshment, go back to this
episode and follow the instructions on creating an R project folder.
Once you have created the project, open the project in RStudio.
Set up your working directory
The working directory is an important concept to understand. It is the place on your computer where R will look for and save files. When you write code for your project, your scripts should refer to files in relation to the root of your working directory and only to files within this structure. Using RStudio projects makes this easy and ensures that your working directory is set up properly.
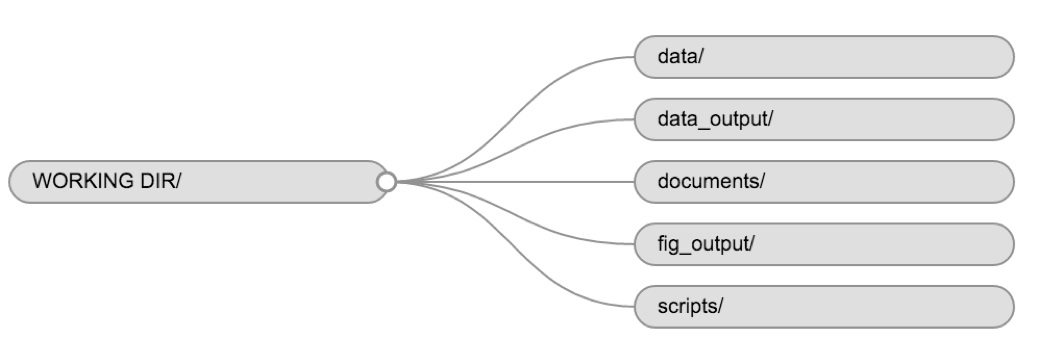
Using a consistent folder structure across your projects will help keep things organized and make it easy to find/file things in the future. This can be especially helpful when you have multiple projects. In general, you might create directories (folders) for scripts, data, and documents. Here are some examples of suggested directories:
-
data/Use this folder to store your raw data and intermediate datasets. For the sake of transparency and provenance, you should always keep a copy of your raw data accessible and do as much of your data cleanup and preprocessing programmatically (i.e., with scripts, rather than manually) as possible. -
data_output/When you need to modify your raw data, it might be useful to store the modified versions of the datasets in a different folder. -
documents/Used for outlines, drafts, and other text. -
fig_output/This folder can store the graphics that are generated by your scripts. -
scripts/A place to keep your R scripts for different analyses or plotting.
You may want additional directories or subdirectories depending on your project needs, but these should form the backbone of your working directory.
Using the getwd() and
setwd() commands
Knowing your current directory is important so that you
save your files, scripts and output in the right location. You can check
your current directory by running getwd() in the RStudio
interface. If for some reason your working directory is not what it
should be, you can change it manually by navigating in the file browser
to where your working directory should be, then by clicking on the blue
gear icon “More”, and selecting “Set As Working Directory”.
Alternatively, you can use
setwd("/path/to/working/directory") to reset your working
directory. However, your scripts should not include this line, because
it will fail on someone else’s computer.
Tips on Using the setwd()
command
Some points to note about setting your working directory:
The directory must be in quotation marks.
For Windows users, directories in file paths are separated with a backslash
\. However, in R, you must use a forward slash/. You can copy and paste from the Windows Explorer window directly into R and use find/replace (Ctrl/Cmd + F) in R Studio to replace all backslashes with forward slashes.For Mac users, open the Finder and navigate to the directory you wish to set as your working directory. Right click on that folder and press the options key on your keyboard. The ‘Copy “Folder Name”’ option will transform into ’Copy “Folder Name” as Pathname. It will copy the path to the folder to the clipboard. You can then paste this into your
setwd()function. You do not need to replace backslashes with forward slashes.
After you set your working directory, you can use ./ to
represent it. So if you have a folder in your directory called
data, you can use read.csv(“./data”) to represent that
sub-directory.
Getting data into R
Downloading the data
Once you have set your working directory, we will create our folder
structure using the dir.create() function.
For this lesson we will use the following folders in our working directory: data/, data_output/ and fig_output/. Let’s write them all in lowercase to be consistent. We can create them using the RStudio interface by clicking on the “New Folder” button in the file pane (bottom right), or directly from R by typing at console:
R
dir.create("data")
dir.create("data_output")
dir.create("fig_output")
To download the dataset, go to the Figshare page for this curriculum
and download the dataset called “books.csv”. The direct
download link is: https://ndownloader.figshare.com/files/22031487.
Place this downloaded file in the data/ directory that you
just created. Alternatively, you can do this directly from R by
copying and pasting this in your terminal (your instructor can place
this chunk of code in the Etherpad):
R
download.file("https://ndownloader.figshare.com/files/22031487",
"data/books.csv", mode = "wb")
Now if you navigate to your data folder, the
books.csv file should be there. We now need to load it into
our R session.
Loading the data into R
R has some base functions for reading a local data file into your R
session–namely read.table() and read.csv(),
but these have some idiosyncrasies that were improved upon in the
readr package, which is installed and loaded with
tidyverse.
R
library(tidyverse) # loads the core tidyverse, including dplyr, readr, ggplot2, purrr
OUTPUT
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ purrr 1.2.1
✔ forcats 1.0.1 ✔ stringr 1.6.0
✔ ggplot2 4.0.1 ✔ tibble 3.3.1
✔ lubridate 1.9.4 ✔ tidyr 1.3.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsMake sure you have the tidyverse package installed in R.
If not, refer back to the episode Before we start on how to
install R packages.
To get our sample data into our R session, we will use the
read_csv() function and assign it to the books
value.
R
books <- read_csv("./data/books.csv")
You will see the message
Parsed with column specification, followed by each column
name and its data type. When you execute read_csv on a data
file, it looks through the first 1000 rows of each column and guesses
the data type for each column as it reads it into R. For example, in
this dataset, it reads SUBJECT as
col_character (character), and TOT.CHKOUT as
col_double. You have the option to specify the data type
for a column manually by using the col_types argument in
read_csv.
You should now have an R object called books in the
Environment pane.
Reading tabular data
read_csv() assumes that fields are delineated by commas,
however, in several countries, the comma is used as a decimal separator
and the semicolon (;) is used as a field delineator. If you want to read
in this type of files in R, you can use the read_csv2
function. It behaves exactly like read_csv but uses
different parameters for the decimal and the field separators. If you
are working with another format, they can be both specified by the user.
Check out the help for read_csv() by typing
?read_csv to learn more. There is also the
read_tsv() for tab-separated data files, and
read_delim() allows you to specify more details about the
structure of your file.
Discussion: Examine the data
Open and examine the data in R. How many observations and variables are there?
The data contains 10,000 observations and 11 variables.
-
CALL...BIBLIO.: Bibliographic call number. Most of these are cataloged with the Library of Congress classification, but there are also items cataloged in the Dewey Decimal System (including fiction and non-fiction), and Superintendent of Documents call numbers. Character. -
X245.ab: The title and remainder of title. Exported from MARC tag 245|ab fields. Separated by a|pipe character. Character. -
X245.c: The author (statement of responsibility). Exported from MARC tag 245|c. Character. -
TOT.CHKOUT: The total number of checkouts. Integer. -
LOUTDATE: The last date the item was checked out. Date. YYYY-MM-DDThh:mmTZD -
SUBJECT: Bibliographic subject in Library of Congress Subject Headings. Separated by a|pipe character. Character. -
ISN: ISBN or ISSN. Exported from MARC field 020|a. Character -
CALL...ITEM: Item call number. Most of these areNAbut there are some secondary call numbers. -
X008.Date.One: Date of publication. Date. YYYY -
BCODE2: Item format. Character. -
BCODE1Sub-collection. Character.
Data frames and tibbles
Data frames are the de facto data structure for tabular data
in R, and what we use for data processing, statistics, and
plotting.
A data frame is the representation of data in the format of a table where the columns are vectors that all have the same length. Because columns are vectors, each column must contain a single type of data (e.g., characters, integers, factors). For example, here is a figure depicting a data frame comprising a numeric, a character, and a logical vector.
A data frame can be created by hand, but most commonly they are
generated by the functions read_csv() or
read_table(); in other words, when importing spreadsheets
from your hard drive (or the web).
A tibble is an extension of R data
frames used by the tidyverse. When the data is read using
read_csv(), it is stored in an object of class
tbl_df, tbl, and data.frame. You
can see the class of an object with class().
Inspecting data frames
When calling a tbl_df object (like books
here), there is already a lot of information about our data frame being
displayed such as the number of rows, the number of columns, the names
of the columns, and as we just saw the class of data stored in each
column. However, there are functions to extract this information from
data frames. Here is a non-exhaustive list of some of these functions.
Let’s try them out!
Size and dimensions
R
dim(books) # returns a vector with the number of rows in the first element,
and the number of columns as the second element (the **dim**ensions of the object)
nrow(books) # returns the number of rows
ncol(books) # returns the number of columnsContent
To examine the contents of a data frame.
R
head(books) # shows the first 6 rows
tail(books) # shows the last 6 rows
Names
R
names(books) # returns the column names (synonym of `colnames()` for
`data.frame` objects)
glimpse(books) # print names of the books data frame to the consoleSummary
R
View(books) # look at the data in the viewer
str(books) # structure of the object and information about the class,
length and content of each column
summary(books) # summary statistics for each columnNote: most of these functions are “generic”, they can be used on other types of objects besides data frames.
The map() function from purrr is a useful
way of running a function on all variables in a data frame or list. If
you loaded the tidyverse at the beginning of the session,
you also loaded purrr. Here we call class() on
books using map_chr(), which will return a
character vector of the classes for each variable.
R
map_chr(books, class)
OUTPUT
CALL...BIBLIO. X245.ab X245.c LOCATION TOT.CHKOUT
"character" "character" "character" "character" "numeric"
LOUTDATE SUBJECT ISN CALL...ITEM. X008.Date.One
"character" "character" "character" "character" "character"
BCODE2 BCODE1
"character" "character" Indexing and subsetting data frames
Our books data frame has 2 dimensions: rows
(observations) and columns (variables). If we want to extract some
specific data from it, we need to specify the “coordinates” we want from
it.
In the last session, we used square brackets [ ] to
subset values from vectors. Here we will do the same thing for data
frames, but we can now add a second dimension. Row numbers come first,
followed by column numbers. However, note that different ways of
specifying these coordinates lead to results with different classes.
R
## first element in the first column of the data frame (as a vector)
books[1, 1]
## first element in the 6th column (as a vector)
books[1, 6]
## first column of the data frame (as a vector)
books[[1]]
## first column of the data frame (as a data.frame)
books[1]
## first three elements in the 7th column (as a vector)
books[1:3, 7]
## the 3rd row of the data frame (as a data.frame)
books[3, ]
## equivalent to head_books <- head(books)
head_books <- books[1:6, ]
Dollar sign
The dollar sign $ is used to distinguish a specific
variable (column, in Excel-speak) in a data frame:
R
head(books$X245.ab) # print the first six book titles
OUTPUT
[1] "Bermuda Triangle /"
[2] "Invaders from outer space :|real-life stories of UFOs /"
[3] "Down Cut Shin Creek :|the pack horse librarians of Kentucky /"
[4] "The Chinese book of animal powers /"
[5] "Judge Judy Sheindlin's Win or lose by how you choose! /"
[6] "Judge Judy Sheindlin's You can't judge a book by its cover :|cool rules for school /"R
# print the mean number of checkouts
mean(books$TOT.CHKOUT)
OUTPUT
[1] 2.2847
unique(), table(), and
duplicated()
Use unique() to see all the distinct values in a
variable:
R
unique(books$BCODE2)
OUTPUT
[1] "a" "w" "s" "m" "e" "4" "k" "5" "n" "o"Take one step further with table() to get quick
frequency counts on a variable:
R
table(books$BCODE2) # frequency counts on a variable
OUTPUT
4 5 a e k m n o s w
1 3 6983 68 3 109 2 21 1988 822 You can combine table() with relational operators:
R
table(books$TOT.CHKOUT > 50) # how many books have 50 or more checkouts?
OUTPUT
FALSE TRUE
9991 9 duplicated() will give you the a logical vector of
duplicated values.
R
duplicated(books$ISN) # a TRUE/FALSE vector of duplicated values in the ISN column
!duplicated(books$ISN) # you can put an exclamation mark before it to get non-duplicated values
table(duplicated(books$ISN)) # run a table of duplicated values
which(duplicated(books$ISN)) # get row numbers of duplicated values
Explore missing values
You may also need to know the number of missing values:
R
sum(is.na(books)) # How many total missing values?
OUTPUT
[1] 14509R
colSums(is.na(books)) # Total missing values per column
OUTPUT
CALL...BIBLIO. X245.ab X245.c LOCATION TOT.CHKOUT
561 12 2801 0 0
LOUTDATE SUBJECT ISN CALL...ITEM. X008.Date.One
0 63 2934 7980 158
BCODE2 BCODE1
0 0 R
table(is.na(books$ISN)) # use table() and is.na() in combination
OUTPUT
FALSE TRUE
7066 2934 R
booksNoNA <- na.omit(books) # Return only observations that have no missing values
Recall how we use na.rm, is.na(),
na.omit(), and complete.cases() when dealing
with vectors.
Exercise
Call
View(books)to examine the data frame. Use the small arrow buttons in the variable name to sort tot_chkout by the highest checkouts. What item has the most checkouts?What is the class of the TOT.CHKOUT variable?
Use
table()andis.na()to find out how many NA values are in the ISN variable.Call
summary(books$TOT.CHKOUT). What can we infer when we compare the mean, median, and max?hist()will print a rudimentary histogram, which displays frequency counts. Callhist(books$TOT.CHKOUT). What is this telling us?
Highest checkouts:
Click, clack, moo : cows that type.class(books$TOT.CHKOUT)returnsnumerictable(is.na(books$ISN))returns 2934TRUEvaluesThe median is 0, indicating that, consistent with all book circulation I have seen, the majority of items have 0 checkouts.
As we saw in
summary(), the majority of items have a small number of checkouts
Logical tests
R contains a number of operators you can use to compare values. Use
help(Comparison) to read the R help file.
| operator | function |
|---|---|
< |
Less Than |
> |
Greater Than |
== |
Equal To |
<= |
Less Than or Equal To |
>= |
Greater Than or Equal To |
!= |
Not Equal To |
%in% |
Has a Match In |
is.na() |
Is NA |
!is.na() |
Is Not NA |
Note that the two equal signs (==) are
used for evaluating equality because one equals sign (=) is
used for assigning variables.
A simple logical test using numeric comparison:
R
1 < 2
OUTPUT
[1] TRUER
1 > 2
OUTPUT
[1] FALSESometimes you need to do multiple logical tests (think Boolean
logic). Use help(Logic) to read the help file.
| operator | function |
|---|---|
& |
boolean AND |
| ` | ` |
! |
Boolean NOT |
any() |
Are some values true? |
all() |
Are all values true? |
Logical Subsetting
We can use logical operators to subset our data, just like how we use
the square brackets [] for subsetting.
For instance, if we want to extract rows with Total Checkout Number of more than 5:
R
books[books$TOT.CHKOUT > 5, ]
Compare the output of the following codes to the previous one:
R
books$TOT.CHKOUT > 5
books[books$TOT.CHKOUT > 5]
What differences did you see in the output?
- Use
getwd()andsetwd()to navigate between directories. - Use
read_csv()from tidyverse to read tabular data into R. - Data frames are made up of vectors of equal length, with each vector representing each column of the data frame.
- Summarise the dimension, content and variables in a data frame.
- Using the square brackets
[]and logical operators to subset data frames.
Content from Data cleaning & transformation with dplyr
Last updated on 2026-02-03 | Edit this page
Estimated time: 80 minutes
Overview
Questions
- How can I create new columns or remove exisitng columns from a data frame?
- How can I rename variables or assign new values to a variable?
- How can I combine multiple commands into a single command?
- How can I export my data frame to a csv file?
Objectives
- Describe the functions available in the
dplyrandtidyrpackages. - Recognise and use the following functions:
select(),filter(),rename(),recode(),mutate()andarrange(). - Combine one or more functions using the ‘pipe’ operator
%>%. - Use the split-apply-combine concept for data analysis.
- Export a data frame to a csv file.
Transforming data with dplyr
We are now entering the data cleaning and transforming phase. While
it is possible to do much of the following using Base R functions (in
other words, without loading an external package) dplyr
makes it much easier. Like many of the most useful R packages,
dplyr was developed by data scientist http://hadley.nz/.
dplyr is a package for making tabular data manipulation
easier by using a limited set of functions that can be combined to
extract and summarize insights from your data. It pairs nicely with
tidyr which enables you to swiftly convert
between different data formats (long vs. wide) for plotting and
analysis.
dplyr is also part of the tidyverse. Let’s
make sure we are all on the same page by loading the
tidyverse and the books dataset we downloaded
earlier.
We’re going to learn some of the most common
dplyr functions:
-
rename(): rename columns -
recode(): recode values in a column -
select(): subset columns -
filter(): subset rows on conditions -
mutate(): create new columns by using information from other columns -
group_by()andsummarize(): create summary statistics on grouped data -
arrange(): sort results -
count(): count discrete values
Getting set up
Make sure you have your Rproj and directories set up from the previous episode.
If you have not completed that step, run the following in your Rproj:
R
library(fs) # https://fs.r-lib.org/. fs is a cross-platform, uniform interface to file system operations via R.
dir_create("data")
dir_create("data_output")
dir_create("fig_output")
download.file("https://ndownloader.figshare.com/files/22031487",
"data/books.csv", mode = "wb")
Then load the tidyverse package
R
library(tidyverse)
OUTPUT
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ purrr 1.2.1
✔ forcats 1.0.1 ✔ stringr 1.6.0
✔ ggplot2 4.0.1 ✔ tibble 3.3.1
✔ lubridate 1.9.4 ✔ tidyr 1.3.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsand the books data we saved in the previous lesson.
R
books <- read_csv("data/books.csv") # load the data and assign it to books
Renaming variables
It is often necessary to rename variables to make them more
meaningful. If you print the names of the sample books
dataset you can see that some of the vector names are not particularly
helpful:
R
glimpse(books) # print names of the books data frame to the console
OUTPUT
Rows: 10,000
Columns: 12
$ CALL...BIBLIO. <chr> "001.94 Don 2000", "001.942 Bro 1999", "027.073 App 200…
$ X245.ab <chr> "Bermuda Triangle /", "Invaders from outer space :|real…
$ X245.c <chr> "written by Andrew Donkin.", "written by Philip Brooks.…
$ LOCATION <chr> "juv", "juv", "juv", "juv", "juv", "juv", "juv", "juv",…
$ TOT.CHKOUT <dbl> 6, 2, 3, 6, 7, 6, 4, 2, 4, 13, 6, 7, 3, 22, 2, 9, 4, 8,…
$ LOUTDATE <chr> "11-21-2013 9:44", "02-07-2004 15:29", "10-16-2007 10:5…
$ SUBJECT <chr> "Readers (Elementary)|Bermuda Triangle -- Juvenile lite…
$ ISN <chr> "0789454165 (hbk.)~0789454157 (pbk.)", "0789439999 (har…
$ CALL...ITEM. <chr> "001.94 Don 2000", "001.942 Bro 1999", "027.073 App 200…
$ X008.Date.One <chr> "2000", "1999", "2001", "1999", "2000", "2001", "2001",…
$ BCODE2 <chr> "a", "a", "a", "a", "a", "a", "a", "a", "a", "a", "a", …
$ BCODE1 <chr> "j", "j", "j", "j", "j", "j", "j", "j", "j", "j", "j", …There are many ways to rename variables in R, but the
rename() function in the dplyr package is the
easiest and most straightforward. The new variable name comes first. See
help(rename).
Here we rename the X245.ab variable. Make sure you assign the output
to your books value, otherwise it will just print it to the
console. In other words, we are overwriting the previous
books value with the new one, with X245.ab
renamed to title.
R
# rename the . Make sure you return (<-) the output to your
# variable, otherwise it will just print it to the console
books <- rename(books,
title = X245.ab)
Tip:
When using the rename() function, the new variable name
comes first.
Side note:
Where does X245.ab come from? That is the MARC field
245|ab. However, because R variables cannot start with a number, R
automatically inserted an X, and because pipes | are not allowed in
variable names, R replaced it with a period.
You can also rename multiple variables at once by running:
R
# rename multiple variables at once
books <- rename(books,
author = X245.c,
callnumber = CALL...BIBLIO.,
isbn = ISN,
pubyear = X008.Date.One,
subCollection = BCODE1,
format = BCODE2,
location = LOCATION,
tot_chkout = TOT.CHKOUT,
loutdate = LOUTDATE,
subject = SUBJECT)
books
OUTPUT
# A tibble: 10,000 × 12
callnumber title author location tot_chkout loutdate subject isbn
<chr> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
1 001.94 Don 2000 Bermuda … writt… juv 6 11-21-2… Reader… 0789…
2 001.942 Bro 1999 Invaders… writt… juv 2 02-07-2… Reader… 0789…
3 027.073 App 2001 Down Cut… by Ka… juv 3 10-16-2… Packho… 0060…
4 133.5 Hua 1999 The Chin… by Ch… juv 6 11-22-2… Astrol… 0060…
5 170 She 2000 Judge Ju… illus… juv 7 04-10-2… Childr… 0060…
6 170.44 She 2001 Judge Ju… illus… juv 6 11-12-2… Conduc… 0060…
7 220.9505 Gil 2001 A young … retol… juv 4 12-01-2… Bible … 0060…
8 225.9505 McC 1999 God's Ki… retol… juv 2 08-06-2… Bible … 0689…
9 292.13 McC 2001 Roman my… retol… juv 4 04-03-2… Mythol… 0689…
10 292.211 McC 1998 Greek go… retol… juv 13 11-16-2… Gods, … 0689…
# ℹ 9,990 more rows
# ℹ 4 more variables: CALL...ITEM. <chr>, pubyear <chr>, format <chr>,
# subCollection <chr>Rename CALL...ITEM.
- Use
rename()to rename theCALL...ITEM.column tocallnumber2. Remember to add the period to the end of theCALL...ITEM.value
R
books <- rename(books,
callnumber2 = CALL...ITEM.)
Recoding values
It is often necessary to recode or reclassify values in your data.
For example, in the sample dataset provided to you, the
sub_collection (formerly BCODE1) and
format (formerly BCODE2) variables contain
single characters.
You can reassign names to the values easily using the
recode() function from the dplyr package.
Unlike rename(), the old value comes first here. Also
notice that we are overwriting the books$subCollection
variable.
R
# first print to the console all of the unique values you will need to recode
distinct(books, subCollection)
OUTPUT
FALSE # A tibble: 10 × 1
FALSE subCollection
FALSE <chr>
FALSE 1 j
FALSE 2 b
FALSE 3 u
FALSE 4 r
FALSE 5 -
FALSE 6 s
FALSE 7 c
FALSE 8 z
FALSE 9 a
FALSE 10 tR
books$subCollection <- recode(books$subCollection,
"-" = "general collection",
u = "government documents",
r = "reference",
b = "k-12 materials",
j = "juvenile",
s = "special collections",
c = "computer files",
t = "theses",
a = "archives",
z = "reserves")
books
OUTPUT
FALSE # A tibble: 10,000 × 12
FALSE callnumber title author location tot_chkout loutdate subject isbn
FALSE <chr> <chr> <chr> <chr> <dbl> <chr> <chr> <chr>
FALSE 1 001.94 Don 2000 Bermuda … writt… juv 6 11-21-2… Reader… 0789…
FALSE 2 001.942 Bro 1999 Invaders… writt… juv 2 02-07-2… Reader… 0789…
FALSE 3 027.073 App 2001 Down Cut… by Ka… juv 3 10-16-2… Packho… 0060…
FALSE 4 133.5 Hua 1999 The Chin… by Ch… juv 6 11-22-2… Astrol… 0060…
FALSE 5 170 She 2000 Judge Ju… illus… juv 7 04-10-2… Childr… 0060…
FALSE 6 170.44 She 2001 Judge Ju… illus… juv 6 11-12-2… Conduc… 0060…
FALSE 7 220.9505 Gil 2001 A young … retol… juv 4 12-01-2… Bible … 0060…
FALSE 8 225.9505 McC 1999 God's Ki… retol… juv 2 08-06-2… Bible … 0689…
FALSE 9 292.13 McC 2001 Roman my… retol… juv 4 04-03-2… Mythol… 0689…
FALSE 10 292.211 McC 1998 Greek go… retol… juv 13 11-16-2… Gods, … 0689…
FALSE # ℹ 9,990 more rows
FALSE # ℹ 4 more variables: callnumber2 <chr>, pubyear <chr>, format <chr>,
FALSE # subCollection <chr>Do the same for the format column. Note that you must
put "5" and "4" into quotation marks for the
function to operate correctly.
R
books$format <- recode(books$format,
a = "book",
e = "serial",
w = "microform",
s = "e-gov doc",
o = "map",
n = "database",
k = "cd-rom",
m = "image",
"5" = "kit/object",
"4" = "online video")
Once you have finished recoding the values for the two variables, examine the dataset again and check to see if you have different values this time:
R
distinct(books, subCollection, format)
Subsetting dataframes
Subset rows with filter()
In the last lesson we learned how to subset a data frame using
brackets. As with other R functions, the dplyr package
makes it much more straightforward, using the filter()
function.
Here we will create a subset of books called
booksOnly, which includes only those items where the format
is books. Notice that we use two equal signs == as the
logical operator:
R
booksOnly <- filter(books, format == "book") # filter books to return only those items where the format is books
You can also use multiple filter conditions. Here, the order matters: first we filter to include only books, then of the results, we include only items that have more than zero checkouts.
R
bookCheckouts <- filter(books,
format == "book",
tot_chkout > 0)
How many items were removed? You can find out functionally with:
R
nrow(books) - nrow(bookCheckouts)
OUTPUT
FALSE [1] 5733You can then check the summary statistics of checkouts for books with more than zero checkouts. Notice how different these numbers are from the previous lesson, when we kept zero in. The median is now 3 and the mean is 5.
R
summary(bookCheckouts$tot_chkout)
OUTPUT
FALSE Min. 1st Qu. Median Mean 3rd Qu. Max.
FALSE 1.000 2.000 3.000 5.281 6.000 113.000If you want to filter on multiple conditions within the same
variable, use the %in% operator combined with a vector of
all the values you wish to include within c(). For example,
you may want to include only items in the format serial and
microform:
R
serial_microform <- filter(books, format %in% c("serial", "microform"))
Exercise: Subsetting with
filter()
Use
filter()to create a data frame calledbooksJuvconsisting offormatbooks andsubCollectionjuvenile materials.Use
mean()to check the average number of checkouts for thebooksJuvdata frame.
R
booksJuv <- filter(books,
format == "book",
subCollection == "juvenile")
mean(booksJuv$tot_chkout)
OUTPUT
[1] 10.41404
Subset columns with select()
The select() function allows you to keep or remove
specific columns It also provides a convenient way to reorder
variables.
R
# specify the variables you want to keep by name
booksTitleCheckouts <- select(books, title, tot_chkout)
booksTitleCheckouts
OUTPUT
# A tibble: 10,000 × 2
title tot_chkout
<chr> <dbl>
1 Bermuda Triangle / 6
2 Invaders from outer space :|real-life stories of UFOs / 2
3 Down Cut Shin Creek :|the pack horse librarians of Kentucky / 3
4 The Chinese book of animal powers / 6
5 Judge Judy Sheindlin's Win or lose by how you choose! / 7
6 Judge Judy Sheindlin's You can't judge a book by its cover :|cool… 6
7 A young child's Bible / 4
8 God's Kingdom :|stories from the New Testament / 2
9 Roman myths / 4
10 Greek gods and goddesses / 13
# ℹ 9,990 more rowsR
# specify the variables you want to remove with a -
books <- select(books, -location)
# reorder columns, combined with everything()
booksReordered <- select(books, title, tot_chkout, loutdate, everything())
Tips:
If your variable name has spaces, use `` to close the variable:
R
select(df, -`my variable`)
ERROR
Error in `UseMethod()`:
! no applicable method for 'select' applied to an object of class "function"Ordering data with arrange()
The arrange() function in the dplyr package
allows you to sort your data by alphabetical or numerical order.
R
booksTitleArrange <- arrange(books, title)
# use desc() to sort a variable in descending order
booksHighestChkout <- arrange(books, desc(tot_chkout))
booksHighestChkout
OUTPUT
# A tibble: 10,000 × 11
callnumber title author tot_chkout loutdate subject isbn callnumber2 pubyear
<chr> <chr> <chr> <dbl> <chr> <chr> <chr> <chr> <chr>
1 E Cro 2000 Clic… by Do… 113 01-23-2… Cows -… 0689… E Cro 2000 2000
2 PZ7.W6367… The … by Da… 106 03-07-2… Pigs -… 0618… 398.2452 W… 2001
3 <NA> Cook… Janet… 103 03-13-2… Cake -… 0152… E Ste 1999 1999
4 PZ7.D5455… Beca… Kate … 79 03-27-2… Dogs -… 0763… Fic Dic 20… 2000
5 PZ7.C6775… Upto… Bryan… 69 02-05-2… Harlem… 9780… E Col 2000 2000
6 <NA> <NA> <NA> 64 08-23-2… <NA> <NA> #1 ENC. C… <NA>
7 F379.N59 … Thro… Ruby … 63 11-01-2… Bridge… 0590… 920 Bri 19… 1999
8 PZ7.C9413… Bud,… Chris… 63 04-03-2… Runawa… 0385… Fic Cur 19… 1999
9 E Mar 1992 Brow… by Bi… 61 02-16-2… Color … 0805… E Mar 1992 1992
10 PZ7.P338 … A ye… Richa… 47 03-26-2… Grandm… 0803… Fic Pec 20… 2000
# ℹ 9,990 more rows
# ℹ 2 more variables: format <chr>, subCollection <chr>R
# order data based on multiple variables (e.g. sort first by checkout, then by publication year)
booksChkoutYear <- arrange(books, desc(tot_chkout), desc(pubyear))
Creating new variables with mutate()
The mutate() function allows you to create new
variables. Here, we use the str_sub() function from the
stringr package to extract the first character of the
callnumber variable (the call number class) and put it into
a new column called call_class.
R
booksLC <- mutate(books,
call_class = str_sub(callnumber, 1, 1))
There are two numbers because you must specify a start and an end value–here, we start with the first character, and end with the first character.
mutate() is also helpful to coerce a column from one
data type to another. For example, we can see there are some errors in
the pubyear variable–some dates are 19zz or
uuuu. As a result, this variable was read in as a
character rather than an integer.
R
books <- mutate(books, pubyear = as.integer(pubyear))
WARNING
Warning: There was 1 warning in `mutate()`.
ℹ In argument: `pubyear = as.integer(pubyear)`.
Caused by warning:
! NAs introduced by coercionWe see the error message NAs introduced by coercion.
This is because non-numerical variables become NA and the
remainder become integers.
Putting it all together with %>%
The Pipe
Operator %>% is loaded with the
tidyverse. It takes the output of one statement and makes
it the input of the next statement. You can think of it as “then” in
natural language. So instead of making a bunch of intermediate data
frames and cluttering up your workspace, you can run multiple functions
at once. You can type the pipe with Ctrl + Shift +
M if you have a PC or Cmd + Shift +
M if you have a Mac.
So in the following example, the books tibble is first
called, then the format is filtered to include only
book, then only the title and
tot_chkout columns are selected, and finally the data is
rearranged from most to least checkouts.
R
myBooks <- books %>%
filter(format == "book") %>%
select(title, tot_chkout) %>%
arrange(desc(tot_chkout))
myBooks
OUTPUT
# A tibble: 6,983 × 2
title tot_chkout
<chr> <dbl>
1 Click, clack, moo :|cows that type / 113
2 The three pigs / 106
3 Cook-a-doodle-doo! / 103
4 Because of Winn-Dixie / 79
5 Uptown / 69
6 Through my eyes / 63
7 Bud, not Buddy / 63
8 Brown bear, brown bear, what do you see? / 61
9 A year down yonder / 47
10 Wemberly worried / 43
# ℹ 6,973 more rowsExercise: Playing with pipes
%>%
- Create a new data frame
booksKidswith these conditions:
-
filter()to includesubCollectionjuvenile & k-12 materials andformatbooks. -
select()only title, call number, total checkouts, and publication year -
arrange()by total checkouts in descending order
- Use
mean()to check the average number of checkouts for thebooksKidsdata frame.
R
booksKids <- books %>%
filter(subCollection %in% c("juvenile", "k-12 materials"),
format == "book") %>%
select(title, callnumber, tot_chkout, pubyear) %>%
arrange(desc(tot_chkout))
mean(booksKids$tot_chkout)
OUTPUT
[1] 9.336331Split-apply-combine and the summarize() function
Many data analysis tasks can be approached using the
split-apply-combine paradigm: split the data into groups, apply
some analysis to each group, and then combine the results.
dplyr makes this very easy through the use
of the group_by() function.
The summarize() function
group_by() is often used together with
summarize(), which collapses each group into a single-row
summary of that group. group_by() takes as arguments the
column names that contain the categorical variables for
which you want to calculate the summary statistics.
So to compute the average checkouts by format:
R
books %>%
group_by(format) %>%
summarize(mean_checkouts = mean(tot_chkout))
OUTPUT
# A tibble: 10 × 2
format mean_checkouts
<chr> <dbl>
1 book 3.23
2 cd-rom 0.333
3 database 0
4 e-gov doc 0.0402
5 image 0.0275
6 kit/object 1.33
7 map 10.6
8 microform 0.00122
9 online video 0
10 serial 0 Books and maps have the highest, and as we would expect, databases, online videos, and serials have zero checkouts.
Here is a more complex example:
R
books %>%
filter(format == "book") %>%
mutate(call_class = str_sub(callnumber, 1, 1)) %>%
group_by(call_class) %>%
summarize(count = n(),
sum_tot_chkout = sum(tot_chkout)) %>%
arrange(desc(sum_tot_chkout))
OUTPUT
# A tibble: 34 × 3
call_class count sum_tot_chkout
<chr> <int> <dbl>
1 E 487 3114
2 <NA> 459 3024
3 H 1142 2902
4 P 800 2645
5 F 240 1306
6 Q 333 1305
7 B 426 1233
8 R 193 981
9 L 358 862
10 5 60 838
# ℹ 24 more rowsLet’s break this down step by step:
- First we call the
booksdata frame - We then pipe through
filter()to include only books - We then create a new column with
mutate()calledcall_classby using thestr_sub()function to keep the first character of thecall_numbervariable - We then
group_by()our newly createdcall_classvariable - We then create two summary columns by using
summarize()- take the number
n()of items percall_classand assign it to a column calledcount - take the the
sum()oftot_chkoutpercall_classand assign the result to a column calledsum_tot_chkout
- take the number
- Finally, we arrange
sum_tot_chkoutin descending order, so we can see the class with the most total checkouts. We can see it is theEclass (History of America), followed byNA(items with no call number data), followed byH(Social Sciences) andP(Language and Literature).
Pattern matching
Cleaning text with the stringr package is easier when
you have a basic understanding of ‘regex’, or regular expression pattern
matching. Regex is especially useful for manipulating strings
(alphanumeric data), and is the backbone of search-and-replace
operations in most applications. Pattern matching is common to all
programming languages but regex syntax is often code-language specific.
Below, find an example of using pattern matching to find and replace
data in R:
- Remove the trailing slash in the title column
- Modify the punctuation separating the title from a subtitle
Note: If the final product of this data will be imported into an ILS, you may not want to alter the MARC specific punctuation. All other audiences will appreciate the text normalizing steps.
Read more about matching patterns with regular expressions.
R
books %>%
mutate(title_modified = str_remove(title, "/$")) %>% # remove the trailing slash
mutate(title_modified = str_replace(title_modified, "\\s:\\|", ": ")) %>% # replace ' :|' with ': '
select(title_modified, title)
OUTPUT
# A tibble: 10,000 × 2
title_modified title
<chr> <chr>
1 "Bermuda Triangle " Berm…
2 "Invaders from outer space: real-life stories of UFOs " Inva…
3 "Down Cut Shin Creek: the pack horse librarians of Kentucky " Down…
4 "The Chinese book of animal powers " The …
5 "Judge Judy Sheindlin's Win or lose by how you choose! " Judg…
6 "Judge Judy Sheindlin's You can't judge a book by its cover: cool rule… Judg…
7 "A young child's Bible " A yo…
8 "God's Kingdom: stories from the New Testament " God'…
9 "Roman myths " Roma…
10 "Greek gods and goddesses " Gree…
# ℹ 9,990 more rowsExporting data
Now that you have learned how to use
dplyr to extract information from or
summarize your raw data, you may want to export these new data sets to
share them with your collaborators or for archival.
Similar to the read_csv() function used for reading CSV
files into R, there is a write_csv() function that
generates CSV files from data frames.
We have previously created the directory data_output
which we will use to store our generated dataset. It is good practice to
keep your input and output data in separate folders–the
data folder should only contain the raw, unaltered data,
and should be left alone to make sure we don’t delete or modify it.
In preparation for our next lesson on plotting, we are going to create a version of the dataset with most of the changes we made above. We will first read in the original, then make all the changes with pipes.
R
books_reformatted <- read_csv("./data/books.csv") %>%
rename(title = X245.ab,
author = X245.c,
callnumber = CALL...BIBLIO.,
isbn = ISN,
pubyear = X008.Date.One,
subCollection = BCODE1,
format = BCODE2,
location = LOCATION,
tot_chkout = TOT.CHKOUT,
loutdate = LOUTDATE,
subject = SUBJECT,
callnumber2 = CALL...ITEM.) %>%
mutate(pubyear = as.integer(pubyear),
call_class = str_sub(callnumber, 1, 1),
subCollection = recode(subCollection,
"-" = "general collection",
u = "government documents",
r = "reference",
b = "k-12 materials",
j = "juvenile",
s = "special collections",
c = "computer files",
t = "theses",
a = "archives",
z = "reserves"),
format = recode(format,
a = "book",
e = "serial",
w = "microform",
s = "e-gov doc",
o = "map",
n = "database",
k = "cd-rom",
m = "image",
"5" = "kit/object",
"4" = "online video"))
This chunk of code read the CSV, renamed the variables, used
mutate() in combination with recode() to
recode the format and subCollection values,
used mutate() in combination with as.integer()
to coerce pubyear to integer, and used
mutate() in combination with str_sub to create
the new varable call_class.
We now write it to a CSV and put it in the data/output
sub-directory:
R
write_csv(books_reformatted, "./data_output/books_reformatted.csv")
Help with dplyr
- Read more about
dplyrat https://dplyr.tidyverse.org/. - In your console, after loading
library(dplyr), runvignette("dplyr")to read an extremely helpful explanation of how to use it. - See the http://r4ds.had.co.nz/transform.html in Garrett Grolemund and Hadley Wickham’s book R for Data Science.
- Watch this Data School video: https://www.youtube.com/watch?v=jWjqLW-u3hc
- Use the
dplyrpackage to manipulate dataframes. - Subset data frames using
select()andfilter(). - Rename variables in a data frame using
rename(). - Recode values in a data frame using
recode(). - Use
mutate()to create new variables. - Sort data using
arrange(). - Use
group_by()andsummarize()to work with subsets of data. - Use pipe (
%>%) to combine multiple commands.
Content from Data Visualisation with ggplot2
Last updated on 2026-02-19 | Edit this page
Estimated time: 115 minutes
Overview
Questions
- What are the components of a ggplot?
- How do I create scatterplots, boxplots, and barplots?
- How can I change the aesthetics (ex. colour, transparency) of my plot?
- How can I create multiple plots at once?
Objectives
- Understand the basic syntax of the
ggplotfunction. - Produce scatter plots, boxplots, and time series plots using ggplot.
- Set universal plot settings.
- Describe what faceting is and apply faceting in ggplot.
- Modify the aesthetics of an existing ggplot plot (including axis labels and color).
- Build complex and customized plots from data in a data frame.
Getting set up
Set up your directories and data
If you have not already done so, open your R Project file
(library_carpentry.Rproj) created in the
Before We Start lesson. Ensure that you have the following
sub-directories in your Rproj: data\,
data_output\ and fig_output\.
Run the following code chunk to create the sub-directories and download the dataset.
R
library(fs) # https://fs.r-lib.org/. fs is a cross-platform, uniform interface to file system operations via R.
dir_create("data")
dir_create("data_output")
dir_create("fig_output")
download.file("https://ndownloader.figshare.com/files/22031487",
"data/books.csv", mode = "wb")
download.file("https://ndownloader.figshare.com/files/22051506",
"data_output/books_reformatted.csv", mode = "wb")
Then load the packages required for this lesson.
R
library(tidyverse) # load the core tidyverse
OUTPUT
── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ purrr 1.2.1
✔ forcats 1.0.1 ✔ stringr 1.6.0
✔ ggplot2 4.0.1 ✔ tibble 3.3.1
✔ lubridate 1.9.4 ✔ tidyr 1.3.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsR
library(lubridate) # load lubridate
Thelubridate package is installed with the tidyverse,
but is not one of the core tidyverse packages loaded with
library(tidyverse), so it needs to be explicitly called.
lubridate makes working with dates and times easier in
R.
We also load the books_reformatted data we saved in the
previous lesson. Here, we’ll assign it to books2. You can
create the reformatted data by running the codes
for exporting data or by loading the saved CSV file from previous
episode.
R
books2 <- read_csv("data_output/books_reformatted.csv") # load the data and assign it to books
Plotting with ggplot2
Base R contains a number of functions for quick data visualization
such as plot() for scatter plots, barplot(),
hist() for histograms, and boxplot(). However,
just as data manipulation is easier with dplyr than Base R,
so data visualization is easier with ggplot2 than Base R.
ggplot2 is a “grammar” for
data visualization also created by Hadley Wickham, as an
implementation of Leland Wilkinson’s Grammar of
Graphics.
ggplot is a plotting package that makes it simple to
create complex plots from data stored in a data frame. It provides a
programmatic interface for specifying what variables to plot, how they
are displayed, and general visual properties. Therefore, we only need
minimal changes if the underlying data change or if we decide to change
from a bar plot to a scatterplot. This helps in creating publication
quality plots with minimal amounts of adjustments and tweaking.
Tips on plotting with ggplot2:
ggplot2 functions like data in the ‘long’ format, i.e.,
a column for every dimension, and a row for every observation.
Well-structured data will save you lots of time when making figures with
ggplot2
ggplot graphics are built step by step by adding new elements. Adding layers in this fashion allows for extensive flexibility and customization of plots.
To build a ggplot, we will use the following basic template that can be used for different types of plots:
ggplot(data = <DATA>, mapping = aes(<MAPPINGS>)) + <GEOM_FUNCTION>()- The
dataargument is used to bind the plot to a specific data frame inggplot2. - The
mappingargument defines the variables mapped to various aesthetics of the plot, e.g. the x and y axis. - The
geom_functionargument defines the type of plot, e.g. barplot, scatter plot, boxplot.
ggplot2 versus
ggplot
People are sometimes confused between ggplot2 and
ggplot. The former refers to the name of the package, while
the latter refers to the function that you run.
When you run the ggplot() function, it plots directly to
the Plots tab in the Files, Plots, Packages, and Help pane (lower
right). Alternatively, you can assign a plot to an R object, then call
print()to view the plot in the Files, Plots, Packages, and
Help pane.
Creating your first plot
Let’s create a booksPlot and limit our visualization to
only items in subCollection general collection, juvenile,
and k-12, and filter out items with NA in
call_class. We do this by using the | key on the
keyboard to specify a boolean OR, and use the !is.na()
function to keep only those items that are NOT NA in the
call_class column.
R
# create a new data frame
booksPlot <- books2 %>%
filter(subCollection == "general collection" |
subCollection == "juvenile" |
subCollection == "k-12 materials",
!is.na(call_class))
To start with, we first bind our data to the ggplot()
function using the data argument.
R
ggplot(data = booksPlot) # a blank canvas

After running this code, a blank canvas is being created. Next, we
define a mapping aesthetic with the mapping argument (using
the aesthetic (aes()) function). This defines the variables
to be plotted and specifies how to present them in the graph, e.g. as
x/y positions or characteristics such as size, shape, color, etc.
R
ggplot(data = booksPlot, mapping = aes(x = call_class)) # define the x axis aesthetic
Here we define the x axes, but because we have not yet added any
geoms, we still do not see any data being visualized.
Defining the geoms
Data is visualized with different “geometric shapes” such as bars and
lines; what are called geoms. In your console, type
geom_ and press the tab key to see the geoms included–there
are over 30. For example:
-
geom_point()for scatter plots, dot plots, etc. -
geom_boxplot()for boxplots -
geom_bar()for barplots -
geom_line()for trend lines, time series, etc.
Each geom takes a mapping argument within the
aes() call. This is called the aesthetic mapping
argument. In other words, inside the geom function is an
aes() function, and inside aes() is a mapping
argument specifying how to map the variables inside the visualization.
ggplot then looks for that variable inside the data argument, and plots
it accordingly.
For example, in the below expression, the call_class
variable is being mapped to the x axis in the geometric
shape of a bar. In this example, the y axis (count) is not specified,
nor is it a variable in the original dataset, but is the result of
geom_bar() binning your data inside each call number class and
plotting the bin counts (the number of items falling into each bin).
To add a geom to the plot use the + operator.
R
# add a bar geom and set call_class as the x axis
ggplot(data = booksPlot, mapping = aes(x = call_class)) +
geom_bar()

We can see that there are about 1,500 books in the H class, 1,000 books in the P class, 700 books in the E class, etc. As we shall see, the number of E and F books are deceptively large, as easy books and fiction books both begin with E and are thus lumped into that category, though they are not of the E and F Library of Congress call number classification.
ggplot tips
- Anything you put in the
ggplot()function can be seen by any geom layers that you add (i.e., these are universal plot settings). This includes the x- and y-axis mapping you set up inaes(). - You can also specify mappings for a given geom independently of the
mapping defined globally in the
ggplot()function. - The
+sign used to add new layers must be placed at the end of the line containing the previous layer. If, instead, the+sign is added at the beginning of the line containing the new layer,ggplot2will not add the new layer and will return an error message.
Applying a different geom
This same exact data can be visualized in a couple different ways by
replacing the geom_histogram() function with either
geom_density() (adding a logarithmic x scale) or
geom_freqpoly():
R
# create a density plot
ggplot(data = booksPlot) +
geom_density(aes(x = tot_chkout)) +
scale_y_log10() +
scale_x_log10()
WARNING
Warning in scale_x_log10(): log-10 transformation introduced infinite values.WARNING
Warning: Removed 2348 rows containing non-finite outside the scale range
(`stat_density()`).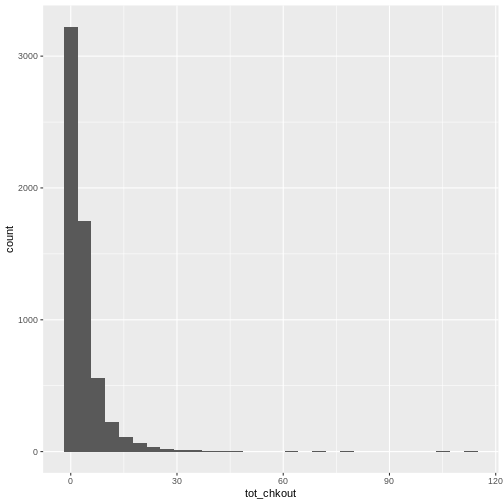
R
# create a frequency polygon
ggplot(data = booksPlot) +
geom_freqpoly(aes(x = tot_chkout), binwidth = 30) +
scale_y_log10()
WARNING
Warning in scale_y_log10(): log-10 transformation introduced infinite values.
Univariate and bivariate geoms
Univariate geoms
“Univariate” refers to a single variable. A histogram is a
univariate plot: it shows the frequency counts of each value inside a
single variable. Let’s say we want to visualize a frequency distibution
of checkouts in the booksPlot data. In other words, how
many items have 1 checkout? How many have 2 checkouts? And so on.
R
ggplot(data = booksPlot, mapping = aes(x = tot_chkout)) +
geom_histogram()
OUTPUT
`stat_bin()` using `bins = 30`. Pick better value `binwidth`.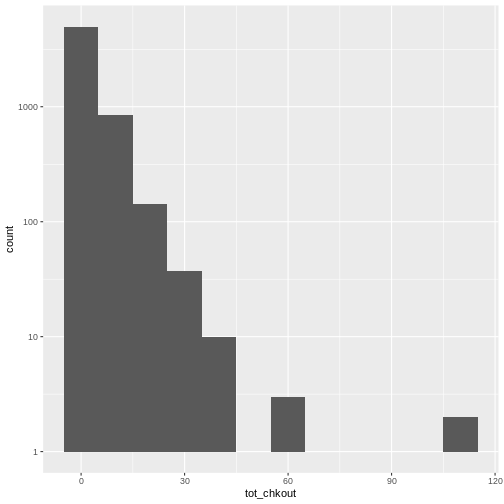
As we have seen in previous lessons, the overwhelming majority of books have a small amount of usage, so the plot is heavily skewed. As anyone who has done collection analysis has encountered, this is a very common issue. It can be addressed in two ways:
First, add a binwidth argument to aes(). In
a histogram, each bin contains the number of occurrences of items in the
data set that are contained within that bin. As stated in the
documentation for ?geom_histogram, “You should always
override this value, exploring multiple widths to find the best to
illustrate the stories in your data.”
Second, change the scales of the y axes by adding
another argument to ggplot:
R
ggplot(data = booksPlot) +
geom_histogram(aes(x = tot_chkout), binwidth = 10) +
scale_y_log10()
WARNING
Warning in scale_y_log10(): log-10 transformation introduced infinite values.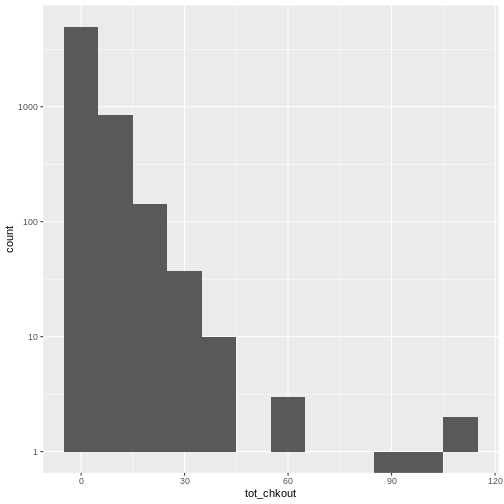
Notice the scale y axis now goes from 0-10, 10-100, 100-1000, and 1000-10000. This is called “logarithmic scale” and is based on orders of magnitude. We can therefore see that over 5,000 books (on the y axis) have between 0-10 checkouts (on the x axis), 1,000 books have 10-20 checkouts, and further down on the x axis, a handful of books have 60-70 checkouts, and a handful more have around 100 checkouts.
We can check this with table():
R
table(booksPlot$tot_chkout)
OUTPUT
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
2348 875 638 464 362 282 199 146 118 97 84 50 41 46 40 33
16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31
17 20 26 17 14 12 7 15 7 8 6 6 3 3 2 4
32 33 34 35 36 38 39 40 41 43 47 61 63 69 79 106
1 5 4 3 2 2 3 1 1 1 1 1 2 1 1 1
113
1 ggplot has thus given us an easy way to visualize the distribution of checkouts. If you test this on your own print and ebook usage data, you will likely find something similar.
Bivariate geoms
Bivariate plots visualize two variables. Let’s take a look at some
higher usage items, but first eliminate the NA values and
keep only items with more than 10 checkouts, which we will do with
filter() from the dplyr package and assign it
to booksHighUsage.
R
# filter booksPlot to include only items with over 10 checkouts
booksHighUsage <- booksPlot %>%
filter(!is.na(tot_chkout),
tot_chkout > 10)
We then visualize checkouts by call number with a scatter plot. There
is still so much skew that I retain the logarithmic scale on the y axis
with scale_y_log10().
R
# scatter plot high usage books by call number class
ggplot(data = booksHighUsage,
aes(x = call_class, y = tot_chkout)) +
geom_point() +
scale_y_log10()
Again, notice the scale on the y axis. We can obseve a few items of
interest here: No items in the D, J, M, and Z class have more than 30
checkouts. An item in the E class has the most checkouts with over 100,
but, as noted above, this includes Easy books classified with
E, not just items with Library of Congress E
classification (United States history) an issue we’ll look at further
down.
Just as with univariate plots, we can use different geoms to view various aspects of the data, which in turn reveal different patterns.
R
# boxplot plot high usage books by call number class
ggplot(data = booksHighUsage,
aes(x = call_class, y = tot_chkout)) +
geom_boxplot() +
scale_y_log10()
By adding points to a boxplot, we can have a better idea of the
number of measurements and of their distribution. Here we set the
boxplot alpha to 0, which will make it
see-through. We also add another layer of geom to the plot called
geom_jitter(), which will introduce a little bit of
randomness into the position of our points. We set the
color of these points to "tomato".
R
ggplot(data = booksHighUsage, aes(x = call_class, y = tot_chkout)) +
geom_boxplot(alpha = 0) +
geom_jitter(alpha = 0.5, color = "tomato") +
scale_y_log10()
Notice how the boxplot layer is behind the jitter layer? What do you need to change in the code to put the boxplot in front of the points such that it’s not hidden?
Plotting Exercise
Boxplots are useful summaries, but hide the shape of the distribution. For example, if the distribution is bimodal, we would not see it in a boxplot. An alternative to the boxplot is the violin plot, where the shape (of the density of points) is drawn.
- Replace the box plot with a violin plot; see
geom_violin().
R
ggplot(data = booksHighUsage, aes(x = call_class, y = tot_chkout)) +
geom_violin(alpha = 0) +
geom_jitter(alpha = 0.5, color = "tomato")
WARNING
Warning: Groups with fewer than two datapoints have been dropped.
ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
Groups with fewer than two datapoints have been dropped.
ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
Groups with fewer than two datapoints have been dropped.
ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
Groups with fewer than two datapoints have been dropped.
ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
Groups with fewer than two datapoints have been dropped.
ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
Plotting Exercise (continued)
- Add color to the
geom_jitterargument on your boxplot according to the itemsubCollection. Hint: If you get the errorobject 'subCollection' not foundorinvalid color name 'subCollection'then considercoloras an aesthetic mapping.
R
ggplot(data = booksHighUsage, aes(x = call_class, y = tot_chkout)) +
geom_violin(alpha = 0) +
geom_jitter(alpha = 0.5, aes(color = subCollection)) +
scale_y_log10()
WARNING
Warning: Groups with fewer than two datapoints have been dropped.
ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
Groups with fewer than two datapoints have been dropped.
ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
Groups with fewer than two datapoints have been dropped.
ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
Groups with fewer than two datapoints have been dropped.
ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
Groups with fewer than two datapoints have been dropped.
ℹ Set `drop = FALSE` to consider such groups for position adjustment purposes.
Plotting Exercise (continued)
So far, we’ve looked at the distribution of checkouts within call number ranges. Try making a new plot to explore the distribution of checkouts within another variable.
- Still using the
booksHighUsagedata, create a boxplot fortot_chkoutfor eachsubCollection. Overlay the boxplot layer on a jitter layer to show actual measurements. Keep thescale_y_log10argument.
R
ggplot(data = booksHighUsage, aes(x = subCollection, y = tot_chkout)) +
geom_boxplot(alpha = 0) +
geom_jitter(alpha = 0.5, color = "tomato") +
scale_y_log10()
Add a third variable
As we saw in that exercise, you can convey even more information in your visualization by adding a third variable, in addition to the first two on the x and y scales.
We can use arguments in aes() to map a visual aesthetic
in the plot to a variable in the dataset. Specifically, we will map
color to the subCollection variable. Because
we are now mapping features of the data to a color, instead of setting
one color for all points, the color now needs to be set inside a call to
the aes function.
ggplot2 will provide a different color
corresponding to different values in the vector. In other words,
associate the name of the aesthetic (color) to the name of
the variable (subCollection) inside aes():
R
ggplot(data = booksHighUsage,
aes(x = call_class,
y = tot_chkout,
color = subCollection)) +
geom_point() +
scale_y_log10()

ggplot() automatically assigns a unique level of the
aesthetic to each unique value of the variable (this is called
scaling). Now we reveal indeed that youth materials make up a
large number of high usage items in both the E and the P class.
Use fill() with geom_bar() to create a
stacked bar plot to visualize frequency. Again, this reinforced the fact
that most of the E and P classification are
youth materials.
R
ggplot(data = booksHighUsage, aes(x = call_class)) +
geom_bar(aes(fill = subCollection))
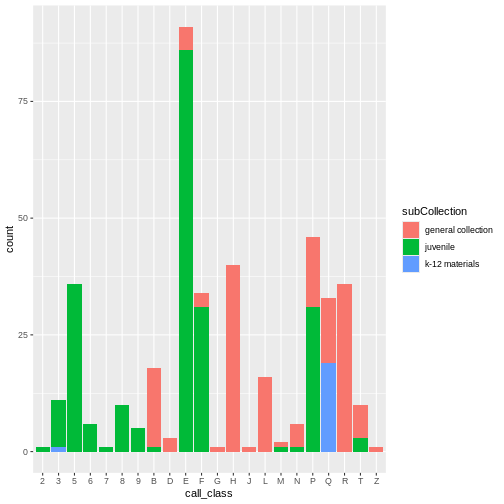
Stacked bar charts are generally more difficult to read than
side-by-side bars. We can separate the portions of the stacked bar that
correspond to each village and put them side-by-side by using the
position argument for geom_bar() and setting
it to “dodge”.
R
ggplot(data = booksHighUsage, aes(x = call_class)) +
geom_bar(aes(fill = subCollection), position = "dodge")
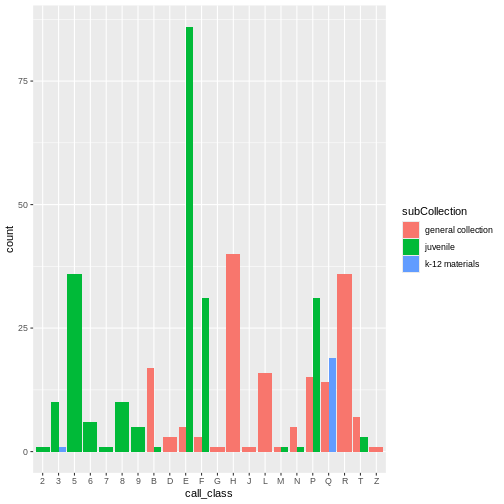
The order of the classification scale is sorted for “library order.”
The audience of library professionals typically prefer an alphabetical
arrangement. However, the x-axis variable is actually categorical.
Categorical data are easier to read when the bars are sorted by
frequency. An easy way to sort by frequency is to use the
fct_infreq() function from the forcats
library.
R
ggplot(data = booksHighUsage, aes(x = fct_infreq(call_class))) +
geom_bar()
Another visualization issue is labeling. In many cultures, long
labels are easier to read horizontally. Our goal is to flip the x-axis
and reorient the x-axis labels into a horizontal presentation. To
accomplish this, flip the axis coordinates with the
coord_flip() function. When we flip the axes it’s important
to reverse the sorted categorical order. Do this with
forcats::fct_rev().
R
ggplot(data = booksHighUsage, aes(x = fct_rev(fct_infreq(call_class)))) +
geom_bar() +
coord_flip()
Plotting time series data
Let’s calculate number of counts per year for each format for items
published after 1990 and before 2002 in the booksHighUsage
data frame created above.
First, we use the ymd() function from the
lubridate package to convert our publication year into a
POSIXct object. Pass the truncated = 2 argument as
a way to indicate that the pubyear column does not contain
month or day. This means 1990 becomes
1990-01-01. This will allow us to plot the number of books
per year.
We will do this by calling mutate() to create a new
variable pubyear_ymd.
R
booksPlot <- booksPlot %>%
mutate(pubyear_ymd = ymd(pubyear, truncated = 2)) # convert pubyear to a Date object with ymd()
class(booksPlot$pubyear) # integer
OUTPUT
[1] "numeric"R
class(booksPlot$pubyear_ymd) # Date
OUTPUT
[1] "Date"Next, we can use filter to remove the NA
values and get books published between 1990 and 2003. Notice that we use
the & as an AND operator to indicate that the date must
fall between that range. We then need to group the data and count
records within each group.
R
yearly_counts <- booksPlot %>%
filter(!is.na(pubyear_ymd),
pubyear_ymd > "1989-01-01" & pubyear_ymd < "2002-01-01") %>%
count(pubyear_ymd, subCollection)
Time series data can be visualized as a line plot with years on the x axis and counts on the y axis:
R
ggplot(data = yearly_counts, mapping = aes(x = pubyear_ymd, y = n)) +
geom_line()
Unfortunately, this does not work because we plotted data for all the
sub-collections together. We need to tell ggplot to draw a line for each
sub-collection by modifying the aesthetic function to include
group = subCollection:
R
ggplot(data = yearly_counts, mapping = aes(x = pubyear_ymd, y = n, group = subCollection)) +
geom_line()
We will be able to distinguish sub-collections in the plot if we add
colors (using color also automatically groups the
data):
R
ggplot(data = yearly_counts, mapping = aes(x = pubyear_ymd, y = n, color = subCollection)) +
geom_line()
Faceting
Rather than creating a single plot with side-by-side bars for each sub-collection, we may want to create multiple plots, where each plot shows the data for a single sub-collection. This would be especially useful if we had a large number of sub-collections that we had sampled, as a large number of side-by-side bars will become more difficult to read.
ggplot2 has a special technique called
faceting that allows the user to split one plot into multiple
plots based on a factor included in the dataset.
There are two types of facet functions:
-
facet_wrap()arranges a one-dimensional sequence of panels to allow them to cleanly fit on one page. -
facet_grid()allows you to form a matrix of rows and columns of panels.
Both geometries allow to to specify faceting variables specified
within vars(). For example,
facet_wrap(facets = vars(facet_variable)) or
facet_grid(rows = vars(row_variable), cols = vars(col_variable)).
Here we use facet_wrap() to make a time series plot for
each subCollection:
R
ggplot(data = yearly_counts, mapping = aes(x = pubyear_ymd, y = n)) +
geom_line() +
facet_wrap(facets = vars(subCollection))
We can use facet_wrap() as a way of seeing the
categories within a variables. Look at the number of formats per
sub-collection.
R
ggplot(data = books2, aes(x = fct_rev(fct_infreq(subCollection)))) +
geom_bar() +
facet_wrap(~ format, nrow = 2) +
scale_y_log10() +
coord_flip() +
labs(x = "Library subcollection", y = "")
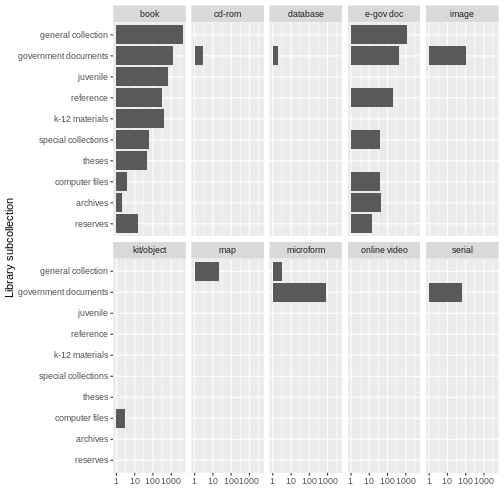
While this may not be the most beautiful plot, these kinds of exercises can be helpful for data exploration. We learn that there are books in all sub-collections; there are CD-ROMS, serials, images, and microforms in government documents, and so on. Exploratory plots visually surface information about your data that are otherwise difficult to parse. Sometimes they may not meet all the rules for creating beautiful data, but when you are simply getting to know your data, that’s OK.
Challenge
Use the books2 data to create a bar plot that depicts
the number of items in each sub-collection, faceted by format. Add the
scale_y_log10() argument to create a logarithmic scale for easier
visibility. Add the following theme argument to tilt the axis text
diagonal:
theme(axis.text.x = element_text(angle = 60, hjust = 1))
R
ggplot(data = books2, aes(x = subCollection)) +
geom_bar() +
facet_wrap(vars(format)) +
scale_y_log10() +
theme(axis.text.x = element_text(angle = 60, hjust = 1))
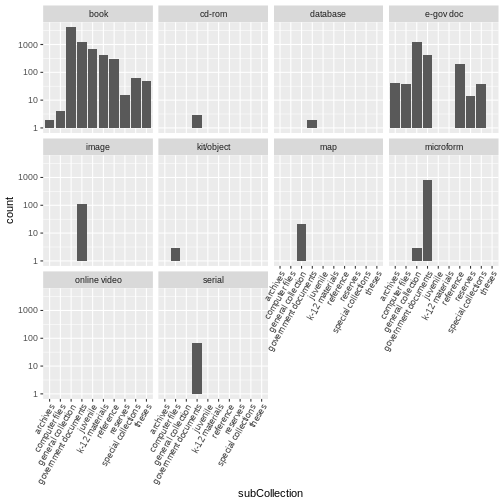
ggplot2 themes
Usually plots with white background look more readable when printed.
Every single component of a ggplot graph can be customized
using the generic theme() function. However, there are
pre-loaded themes available that change the overall appearance of the
graph without much effort.
For example, we can change our graph to have a simpler white
background using the theme_bw() function:
R
#
ggplot(data = yearly_counts, mapping = aes(x = pubyear_ymd, y = n)) +
geom_line() +
facet_wrap(facets = vars(subCollection)) +
theme_bw()
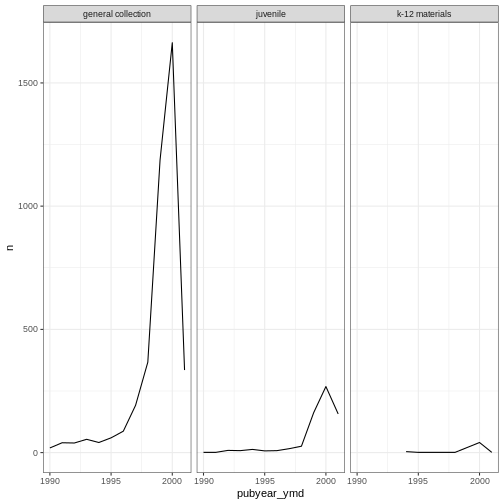
In addition to theme_bw(), which changes the plot
background to white, ggplot2 comes with
several other themes which can be useful to quickly change the look of
your visualization. The complete list of themes is available at https://ggplot2.tidyverse.org/reference/ggtheme.html.
theme_minimal() and theme_light() are popular,
and theme_void() can be useful as a starting point to
create a new hand-crafted theme.
The ggthemes
package provides a wide variety of options. The ggplot2
extensions website provides a list of packages that extend the
capabilities of ggplot2, including
additional themes.
Customization
Take a look at the ggplot2
cheat sheet, and think of ways you could improve your plots.
For example, by default, the axes labels on a plot are determined by the name of the variable being plotted. We can change names of axes to something more informative than ‘pubyear_ymd’ and ‘n’ and add a title to the figure:
R
# add labels
ggplot(data = yearly_counts, mapping = aes(x = pubyear_ymd, y = n)) +
geom_line() +
facet_wrap(facets = vars(subCollection)) +
theme_bw() +
labs(title = "Number of High Usage Books per Year of Publication, by Sub-Collection",
x = "Year of publication",
y = "Number of books")
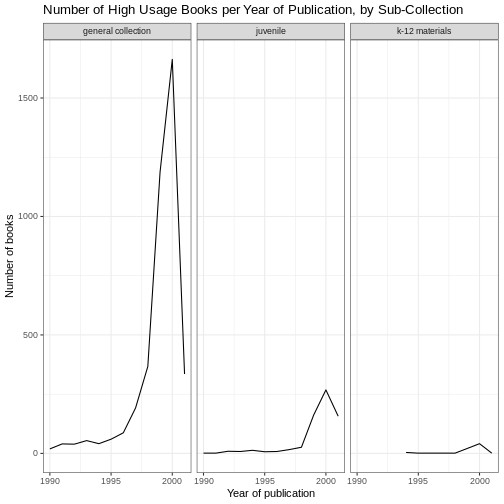
Note that it is also possible to change the fonts of your plots. If
you are on Windows, you may have to install the extrafont
package, and follow the instructions included in the README for this
package.
You can also assign a theme to an object in your
environment, and pass that theme to your plot. This can be helpful to
keep your ggplot() calls less cluttered. Here we create a
gray_theme :
R
# create the gray theme
gray_theme <- theme(axis.text.x = element_text(color = "gray20", size = 12, angle = 45, hjust = 0.5, vjust = 0.5),
axis.text.y = element_text(color = "gray20", size = 12),
text = element_text(size = 16),
plot.title = element_text(hjust = 0.5))
# pass the gray theme to a plot
ggplot(data = yearly_counts, mapping = aes(x = pubyear_ymd, y = n)) +
geom_line() +
facet_wrap(facets = vars(subCollection)) +
gray_theme +
labs(title = "Number of High Usage Books per Year of Publication, \n by Sub-Collection",
x = "Year of publication",
y = "Number of books")
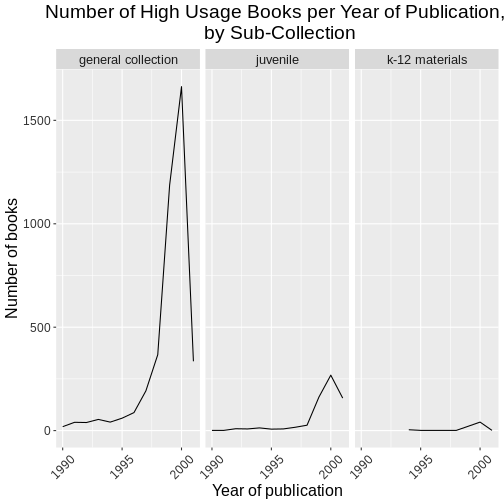
Challenge
Use the booksPlot data to create a plot that depicts how
the total number of checkouts changes based on year of publication.
First, create a data frame yearly_checkouts That meets
the following conditions:
-
filter()to excludeNAvalues -
filter()between “1989-01-01” and “2002-01-01” -
group_by()the publication year (make sure to use the special Date pubyear value we created) -
summarize()to create a new valuecheckouts_sumthat represents thesum()of total checkouts per publication year
Then, create a ggplot that visualizes the sum of item
checkouts by year of publication. Add one of the themes listed
above.
R
yearly_checkouts <- booksPlot %>%
filter(!is.na(pubyear_ymd),
pubyear_ymd > "1989-01-01" & pubyear_ymd < "2002-01-01") %>%
group_by(pubyear_ymd) %>%
summarize(checkouts_sum = sum(tot_chkout))
ggplot(data = yearly_checkouts, mapping = aes(x = pubyear_ymd, y = checkouts_sum)) +
geom_line() +
theme_bw()
Save and export
After creating your plot, you can save it to a file in your favorite format. The Export tab in the Plot pane in RStudio will save your plots at low resolution, which will not be accepted by many journals and will not scale well for posters.
Instead, use the ggsave() function, which allows you
easily change the dimension and resolution of your plot by adjusting the
appropriate arguments (width, height and
dpi). We have been printing our plot output directly to the
console. To use ggsave(), first assign the plot to a
variable in your R environment, such as
yearly_counts_plot.
Make sure you have the fig_output/ folder in your
working directory.
R
yearly_counts_plot <- ggplot(data = yearly_counts, mapping = aes(x = pubyear_ymd, y = n)) +
geom_line() +
facet_wrap(facets = vars(subCollection)) +
gray_theme +
labs(title = "Number of High Usage Books per Year of Publication, \n by Sub-Collection",
x = "Year of publication",
y = "Number of books")
ggsave("fig_output/yearly_counts_plot.png", yearly_counts_plot, width = 15, height = 10)
Exercise
With all of this information in hand, please take another five
minutes to either improve one of the plots generated in this exercise or
create a beautiful graph of your own. Use the RStudio ggplot2
cheat sheet for inspiration. Here are some ideas:
- See if you can make the bars white with black outline.
- Try using a different color palette (see http://www.cookbook-r.com/Graphs/Colors_(ggplot2)/).
-
ggplot2is a flexible and useful tool for creating plots in R. - The data set and coordinate system can be defined using the
ggplotfunction. - Additional layers, including geoms, are added using the
+operator. - Boxplots are useful for visualizing the distribution of a continuous variable.
- Barplot are useful for visualizing categorical data.
- Faceting allows you to generate multiple plots based on a categorical variable.
Content from Reproducible Reports with Quarto
Last updated on 2026-02-19 | Edit this page
Estimated time: 60 minutes
Overview
Questions
- How can I combine my code, results, and narrative into a single document?
- How can I automatically update my reports when my data changes?
- What is Quarto and how does it differ from a standard R script?
Objectives
- Describe the value of reproducible reporting.
- Create a new Quarto document (
.qmd) in RStudio. - Use Markdown syntax to format text.
- Create and run code chunks within a Quarto document.
- Render a Quarto document to an HTML report.
Introduction to Reproducible Reporting
So far, we have been writing code in .R scripts. This is
excellent for data analysis, but what happens when you need to share
your findings with a colleague or a library director? You might copy a
plot into a Word document or an email, then type out your
interpretation.
But what if the data changes next month? You would have to re-run your script, re-save the plot, copy it back into Word, and update your text. This manual process is prone to errors and tedious.
Quarto allows you to combine your code, its output (plots, tables), and your narrative text into a single document. When you “render” the document, R runs the code and produces a polished report (HTML, PDF, or Word) automatically.
Creating a Quarto Document
To create a new Quarto document in RStudio:
- Click the File menu.
- Select New File > Quarto Document…
- In the dialog box, give your document a Title (e.g., “Library Usage Report”) and enter your name as Author.
- Ensure HTML is selected as the output format.
- Click Create.
RStudio will open a new file with some example content. Notice the
file extension is .qmd.
Quarto vs. RMarkdown
If you have used R before, you might be familiar with RMarkdown
(.Rmd). Quarto (.qmd) is the next-generation
version of RMarkdown. It works very similarly but supports more
languages (like Python and Julia) and has better features for scientific
publishing.
Anatomy of a Quarto Document
A Quarto document has three main parts:
1. The YAML Header
At the very top, enclosed between two lines of ---, is
the YAML Header. This contains metadata about the
document.
2. Markdown Text
The white space is where you write your narrative. You use Markdown syntax to format text.
-
**Bold**for bold text -
*Italics*for italics -
# Heading 1for a main title -
## Heading 2for a section title -
- List itemfor bullet points
3. Code Chunks
Code chunks are where your R code lives. They start with
```{r} and end with ```.
``` r
# This is a code chunk
summary(cars)
```
``` output
speed dist
Min. : 4.0 Min. : 2.00
1st Qu.:12.0 1st Qu.: 26.00
Median :15.0 Median : 36.00
Mean :15.4 Mean : 42.98
3rd Qu.:19.0 3rd Qu.: 56.00
Max. :25.0 Max. :120.00
```You can insert a new chunk by clicking the +C button in the editor toolbar, or by pressing Ctrl+Alt+I (Windows/Linux) or Cmd+Option+I (Mac).
Your First Report
Let’s clean up the example file and create a report using our
books data.
- Delete everything in the file below the YAML header.
- Add a new setup code chunk to load our libraries and prepare the data.
```{{r}} #| label: setup #| include: false
library(tidyverse)
Load data and rename columns for clarity
books2 <- read_csv(“data/books.csv”) %>% rename( subCollection = BCODE1, tot_chkout = TOT.CHKOUT, format = BCODE2 ) %>% mutate( subCollection = recode(subCollection, “-” = “general collection”, j = “juvenile”, b = “k-12 materials” ) )
::::::::::::::::::::::::::::::::::::::::: callout
## Chunk Options
Notice the lines starting with `#|`. These are **chunk options**.
- `#| label: setup` gives the chunk a name.
- `#| include: false` runs the code but hides the code and output from the final report. This is great for loading data silently.
:::::::::::::::::::::::::::::::::::::::::
### Adding Analysis
Now, let's add a section header and some text.
```markdown
## High Usage Items
We are analyzing items with more than 10 checkouts to understand circulation patterns across sub-collections.Next, insert a new code chunk and paste the plotting code we developed in the previous episode (ggplot2).
<img src="fig/05-reproducible-reports-rendered-plot-high-usage-1.png" alt="" style="display: block; margin: auto;" />Setting #| echo: false will display the plot in
the report, but hide the R code that generated it. This is
often preferred for reports intended for non-coders.
Rendering the Document
Now comes the magic. Click the Render button (blue arrow icon) at the top of the editor pane.
RStudio will: 1. Run all your code chunks from scratch. 2. Generate
the plots and results. 3. Combine them with your text. 4. Create a new
file named library_usage_report.html in your project
folder. 5. Open a preview of the report.
Challenge: Add a Summary Table
- Add a new header
## Summary Statisticsto your Quarto document. - Insert a new code chunk.
- Write code to calculate the mean checkouts per format (Hint: use
group_by(format)andsummarize()). - Render the document again to see your new table included in the report.
Add this to your document:
``` r
books2 %>%
group_by(format) %>%
summarize(mean_checkouts = mean(tot_chkout, na.rm = TRUE)) %>%
arrange(desc(mean_checkouts))
```
``` output
# A tibble: 10 × 2
format mean_checkouts
<chr> <dbl>
1 map 10.6
2 book 3.23
3 kit/object 1.33
4 cd-rom 0.333
5 e-gov doc 0.0402
6 image 0.0275
7 microform 0.00122
8 database 0
9 online video 0
10 serial 0
```Render the document to see the updated report.
Why This Matters
By using Quarto, your report is now reproducible.
If you download a new version of books.csv next month:
1. Save it to your data/ folder. 2. Open your Quarto
document. 3. Click Render.
Your report will automatically update with the new data, creating a fresh plot and table without you having to copy-paste a single thing.
- Quarto allows you to mix code and text to create reproducible reports.
- Use the YAML header to configure document metadata like title and output format.
-
Code chunks run R code and can display or hide
input/output using options like
#| echo: false. - Rendering the document executes the code and produces the final output (HTML, PDF, etc.).
- This workflow saves time and reduces errors when reporting on data that changes over time.
